- R&D
- Clinical Diagnosis
- News
- Company
QuickBiology News
News Center
Here are relevant industry news and updates on our company
WGS
Whole Genome Sequencing (WGS) has revolutionized the fields of genomics and precision medicine. By enabling the comprehensive analysis of an individual's entire genetic makeupRead More
RNA Sequencing
In the rapidly advancing field of genomics, RNA sequencing (RNA-seq) has become an essential tool for understanding gene expression. Two primary approaches have emerged: single-cell RNA sequencing (scRNA-seq) and bulk RNA sequencing (bulk RNA-seq). Read More
WES
Whole Exome Sequencing (WES) is a powerful and revolutionary technology that has transformed the field of genomics and personalized medicine.Read More
RNA Sequencing
In the age of genomics, RNA sequencing (RNAseq) stands out as a transformative technology, providing deep insights into the transcriptome—the complete set of RNA transcripts produced by the genome under specific circumstances or in a particular cell. Read More
NGS
Next-generation sequencing (NGS) has revolutionized biological and medical research, enabling high-throughput sequencing of DNA and RNA. This technology has facilitated significant advances in genomics, personalized medicine, and molecular diagnostics. Read More
Genomic
Quick Biology Inc. has set up 10X Genomics’ Single Cell Fixed RNA Profiling Sequencing platform.The Single Cell Fixed RNA Profiling (FRP) workflow measures RNA levels in samples (single cells or nuclei) fixed with formaldehyde, using probes targeting the whole transcriptome.Read More
Acute kidney injury (AKI) is a common clinical condition associated with diverse etiologies and abrupt loss of renal function. Read More
Core regulatory circuitry (CRC)-dependent transcriptional network is critical for developmental tumors in children and adolescents carrying few gene mutations. Read More
Genomic
Gastrointestinal adenocarcinomas (GIAC) of the tubular gastrointestinal (GI) tract including esophagus, stomach, colon, and rectum comprise most GI cancers and share a spectrum of genomic features. Read More
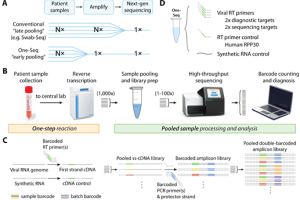
NGS-based SARS-CoV-2 virus detection not only eliminates bottlenecks in scalability but also identifies viral variants, which plays a big role in current SARS-CoV2 VOC (Variants of concerns) surveillance. Previously, Quick Biology reported several scientists’ works, that followed traditional barcoding and sequencing workflows, and required individual RNA extraction and PCR thermocycling steps. In recent medRxiv, researchers from Harvard Medical School reported a new method “One-Seq”, that eliminates the current bottlenecks in scalability by enabling early pooling of samples, before any extraction or amplification steps. Read More