2021-03-29 By Quick Biology
Multiplex detection is the test that numerous analyte targets and internal controls are conducted simultaneously in one reaction. In nucleic acid detection, the gold standard RT-qPCR is multiplexable, which uses the TaqMan method that each target probe is labeled by different fluorescence. The benefits of multiplexing not only reduce reaction costs by at least 50% but also give “self-calibrated” data as all analytes are conducted in the same reaction.
Isothermal amplification of virus detection is a promising technology, which uses fewer instruments, making Point-of-Care or At-Home testing possible. However, all isothermal amplification methods currently such as RPA (recombinase polymerase amplification), LAMP (Loop-mediated isothermal amplification), NDA (nicking endonuclease dependent DNA amplification) have non-specific amplification, their multiplexing capabilities are under-investigated.
In BioTechniques, Dr. Tanner from New England BioLab described the multiplexable RT-LAMP (Reverse-transcription loop-mediated isothermal amplification) method (ref1). Basically, they use Detection of Amplification by Releasing of Quenching (DARQ) that supplements a standard LAMP primer set with a pair of oligonucleotides in duplex form for detection. This duplex consists of a 5’-modified version of the forward inner primer (FIP), Q-FIP, annealed to a 3’-modified oligonucleotide representing the F1 region (Fd), complementary to the FIP in its 5’ section. The two modifications are selected to be dark quencher–fluorophore pairs; when the quenched FIP is incorporated into the LAMP product, subsequent amplification displaces the Fd oligo, releasing quenching and producing a fluorescent signal specific to the label and target of interest (Fig.1, ref2). By DARQ, their multiplexed RT-LAMP detects four targets (SARS-CoV-2, influenza A, influenza B, human RNA), and it is real-time, endpoint fluorescence detection (Fig.2, ref1).
Figure 1: DARQ in LAMP. (A) Schematic depiction of a DARQ probe, with a 5′-quencher FIP (F1c + F2 sequence) annealed to a 3′-fluorophore Fd. The quencher and fluorophore moieties are represented by Q and F, respectively. (B) Outline of DARQ LAMP reactions, with core LAMP primers FIP (F1c + F2), BIP (B1c + B2), F3 and B3, and the DARQ oligonucleotide, Fd (Q, black; F, red). For clarity, LoopF and LoopB primers are not shown. (1) LAMP initiates at the F2c sequence of the target, with the Fd probe quenched through annealing to Q-FIP. This new strand is displaced by upstream synthesis from the F3 primer. (2) The BIP primer anneals to the B2c site in the newly synthesized strand. (3) Synthesis from the primer annealed to the B2c sequence displaces the Fd probe. This releases the quenching resulting in a gain of signal. The newly synthesized strand is displaced by extension from the B3 primer. (4) The resulting structure undergoes exponential amplification in the LAMP reaction. Subsequent initiations at FIP give rise to additional release of Fd, resulting in exponential signal detection. (ref2)
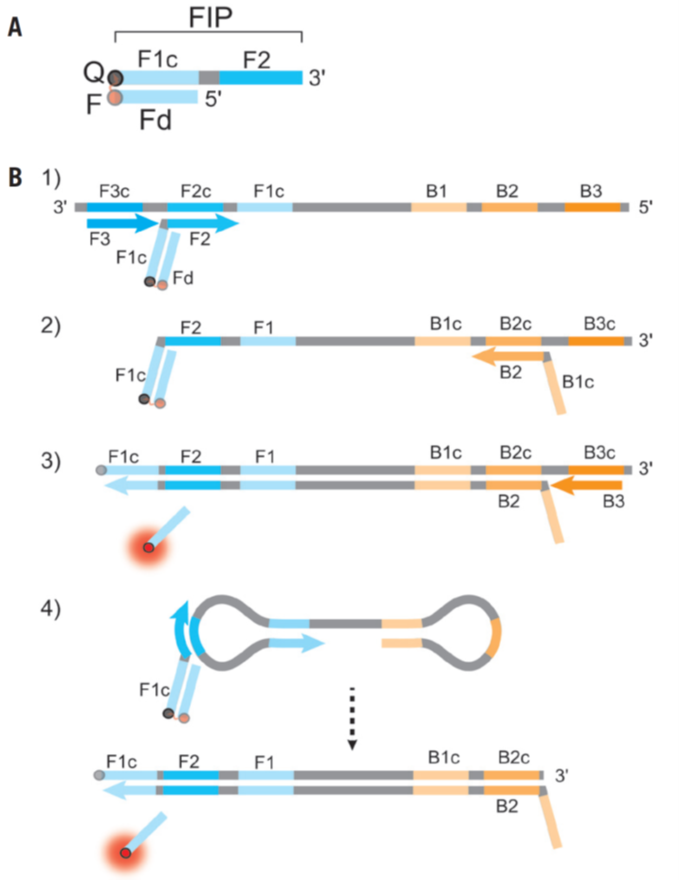
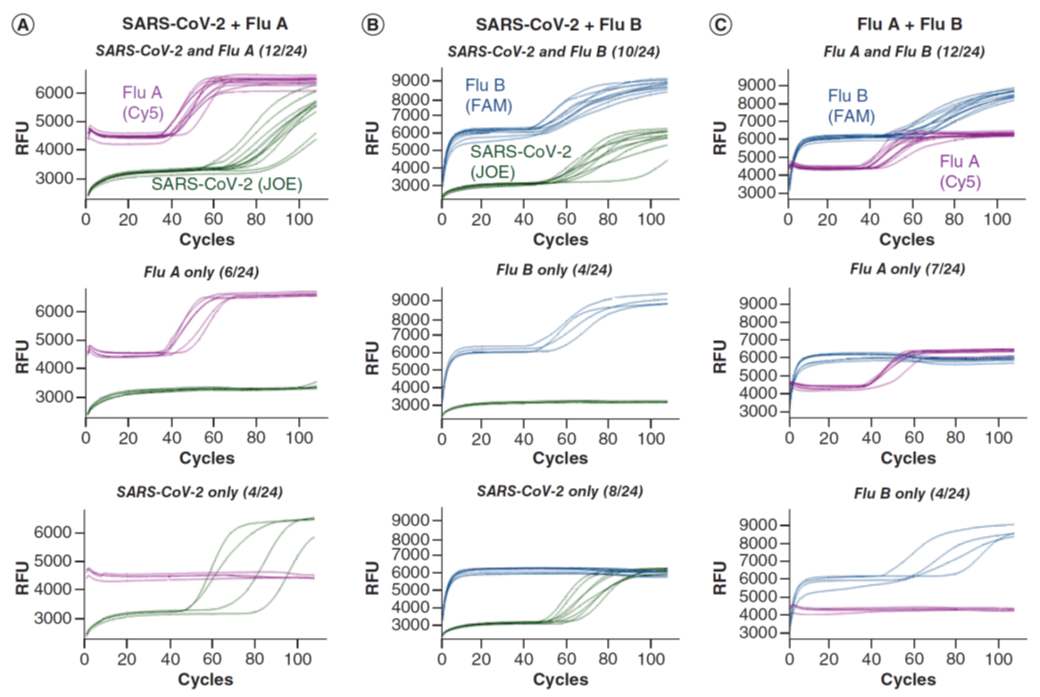
Figure 2: Simultaneous detection of two viral targets and an internal control. Reactions were performed in the presence of E1, IAV, IBV and ACTB primer sets and their QFIP:Fd duplexes with 24 repeats (for simplicity the ROX signal from ACTB is not shown). The targets are pairs of SARS-CoV-2, influenza A and influenza B RNAs. For each target pair, the reactions that had both targets amplified are shown in the top, and reactions that only had one target amplified are shown in the middle and bottom. The fractional numbers in brackets indicate the number of reactions in each category. (A) SARS-CoV-2, influenza A and Jurkat RNAs. (B) SARS-CoV-2, influenza B and Jurkat RNAs. (C) Influenza A, influenza B and Jurkat RNAs. IAV: Influenza A virus primer; IBV: Influenza B virus primer; QFIP: Quencher-modified forward inner primer. (ref1)
Quick Biology can assist you with the SARS-CoV2 viral sequencing project. Find More at Quick Biology.
See resource:
- 1. Zhang, Y. & A Tanner, N. Development of multiplexed reverse-transcription loop-mediated isothermal amplification for detection of SARS-CoV-2 and influenza viral RNA. Biotechniques 70, 167–174 (2021).
- 2. Tanner, N. A., Zhang, Y. & Evans, T. C. Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. Biotechniques 53, 81–89 (2012).



