2021-03-22 By Quick Biology
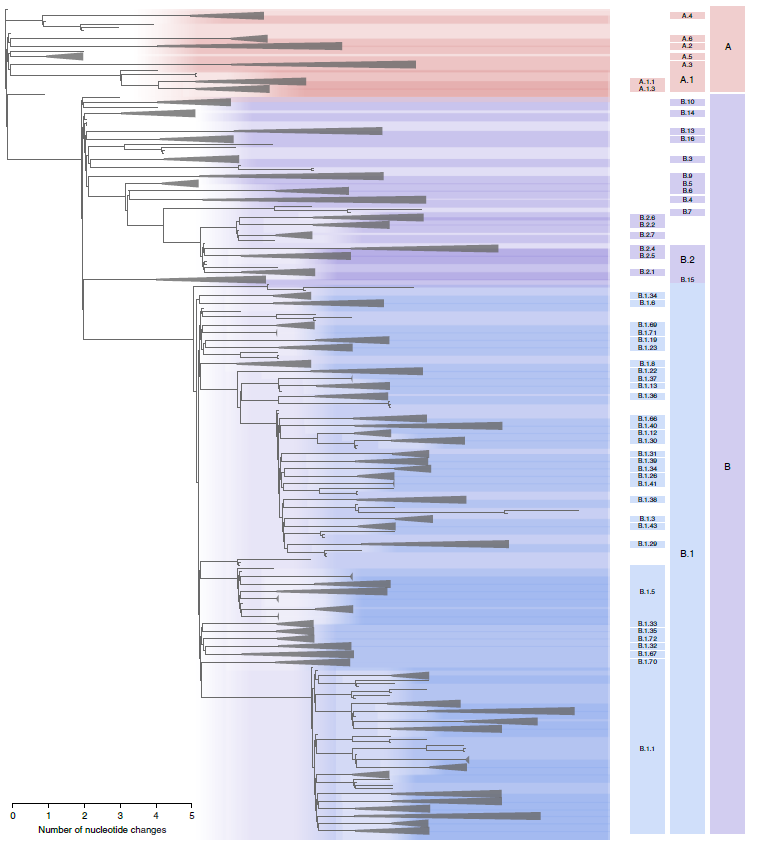
The ongoing pandemic of SARS-CoV-2 has resulted in the generation of hundreds of thousands of virus genome sequences, as of Feb 3, 2021, 468,000 sequences of SARS-CoV-2 from COVID-19 patients have been uploaded into public databases. A rational and dynamic virus nomenclature use a phylogenetic framework to identify lineages. Major lineage labels begin with a letter. Two lineages are at the root of the phylogeny of SARS-CoV-2 as lineage A and lineage B, Wuhan-hu-1 (GenBank accession no. MN908947) sampled on Dec 26 20219 is an early representative, assigned to lineage B. For further lineage designations, a numerical value is assigned, for example, A.1 or B.2 (Fig.1, ref1).
SARS-CoV-2, like other RNA viruses, constantly changes through mutations with new variants occurring over time. Among these variants, a very small proportion are of public health concern because they are more transmissible, cause more severe illness, or can elude the immune response that develops following infection and possibly from vaccination, we name these variants as VOC (variant of concerns) (Ref2).
Since Dec 2020, three VOCs have emerged and merit close monitoring: B.1.1.7/501Y.V1; B.1.351/501Y.V2 and P.1/501Y.V3. CDC (Centers for Disease Control and Prevention) has detailed descriptions of these variants (ref3). Basically, these VOCs happen in either N-terminal or receptor-binding domain (RBD) of the virus Spike gene. Each VOC has been reported with unique mutation profiles, Sanger or next-generation sequencing targeting whole or partial S-gene region is strongly recommended for diagnostic screening and validation. This surveillance is key as VOC is circulating globally, WHO regional office for Europe also provides guidance to laboratories, microbiology experts, and relevant stakeholders in making decisions on establishing or scaling up capability and capacity to detect and identify circulating SARS-CoV-2 variants, and in making decisions on which technologies to use and for which objective (ref4).
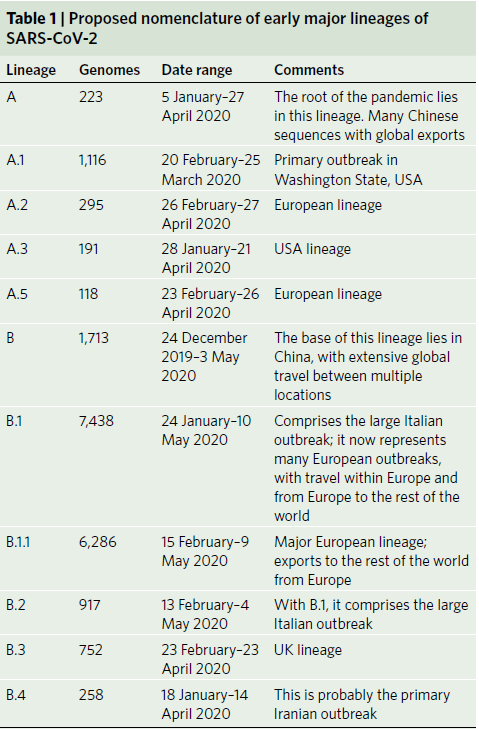
Figure 1: Maximum likelihood phylogeny of globally sampled sequences of SARS-CoV-2 downloaded from the GISAID database on 18 May 2020. Five representative genomes are included from each of the defined lineages. The largest lineages defined by our proposed nomenclature system are highlighted with colored areas and labeled on the right. The remaining lineages defined by the nomenclature system are denoted by triangles. The scale bar represents the number of nucleotide changes within the coding region of the genome. Table 1 summarized the Proposed nomenclature of early major lineages of SARS-CoV-2.
Quick Biology can assist you with SARS-CoV2 WGS. Find More at Quick Biology.
See resource:
- 1. Rambaut, A. et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat. Microbiol. 5, 1403–1407 (2020).
- 2. Walensky, R. P., Walke, H. T. & Fauci, A. S. SARS-CoV-2 Variants of Concern in the United States—Challenges and Opportunities. Jama 30333, 2–3 (2021).
- 3. CDC: https://www.cdc.gov/coronavirus/2019-ncov/more/science-and-research/scientific-brief-emerging-variants.html
- 4. ECDC:https://www.ecdc.europa.eu/en/publications-data/methods-detection-and-identification-sars-cov-2-variants



