2021-03-15 By Quick Biology
Antimicrobial resistance (AMR) is a serious public health concern, especially in Asian countries due to antibiotic overuse. The mechanism of antibiotic resistance is complex but is generally grouped into three broad categories: target modification, drug inactivation, and drug transport.
In the classical approach, evolution experiments of Escherichia coli rely on growth-dependent selection, providing a limited view of the antibiotic resistance landscape (Fig.1, ref1). In recent Science, Prof. James J. Collins’s group designed and implemented an alternative evolution protocol consisting of short-term drug exposure at incrementally increasing metabolic activities, separated by rounds of drug-free growth, to minimize growth-dependent selection and lineage extinction (Fig.2, ref1). By sequencing and analyzing E. coli adapted to representative antibiotics at increasingly heightened metabolic states, they revealed various underappreciated noncanonical genes resulting in metabolic alteration, such as lowering basal respiration, thus avoiding metabolic toxicity and minimizing drug lethality (Fig.3). They also find several metabolism-specific mutations are overrepresented in the genome of > 3500 clinical E.coli pathogens, indicating clinical relevance.
Figure 1: Classic evolution schematic and characterization. Triplicate populations were treated with no antibiotic, strep, carb, or Cipro at increasing concentrations from 0.08× to 40× MIC50 over 11 days (days 0 to 10). MIC50 = Minimum Inhibitory Concentration required to inhibit the growth of 50% of organisms.
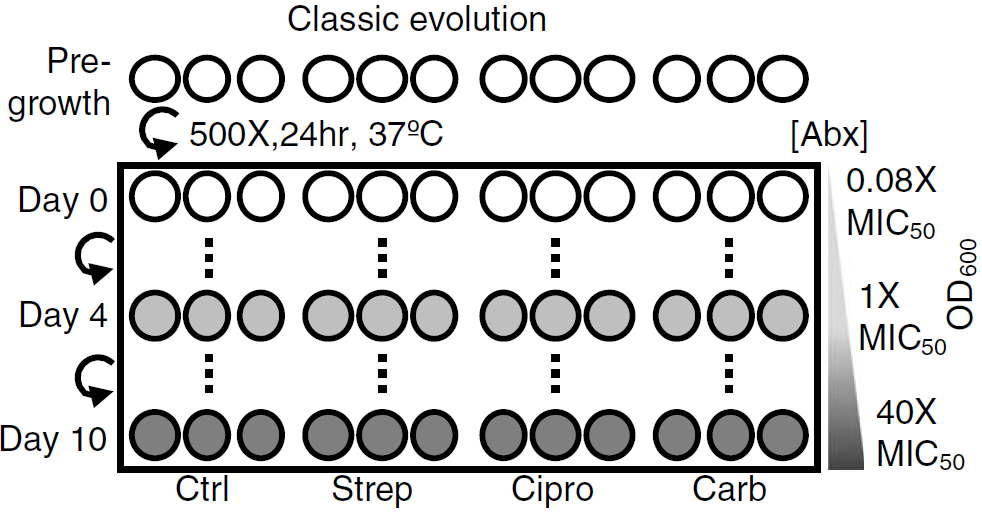
Figure 2: Metabolic evolution leads to acquired resistance with no obvious growth defect. Triplicate populations were treated with no antibiotic, strep, carb, or Cipro for a total of 11 days at 40× MIC50 for 1 hour at increasing metabolic states. Thirty minutes before antibiotic treatment, cells were equilibrated to a temperature that increased daily in 1°C increment beginning on day 0 at 20°C and concluding on day 10 at 30°C. After antibiotic treatment, cells were washed 2× in PBS and grown analogously to the classic evolution.
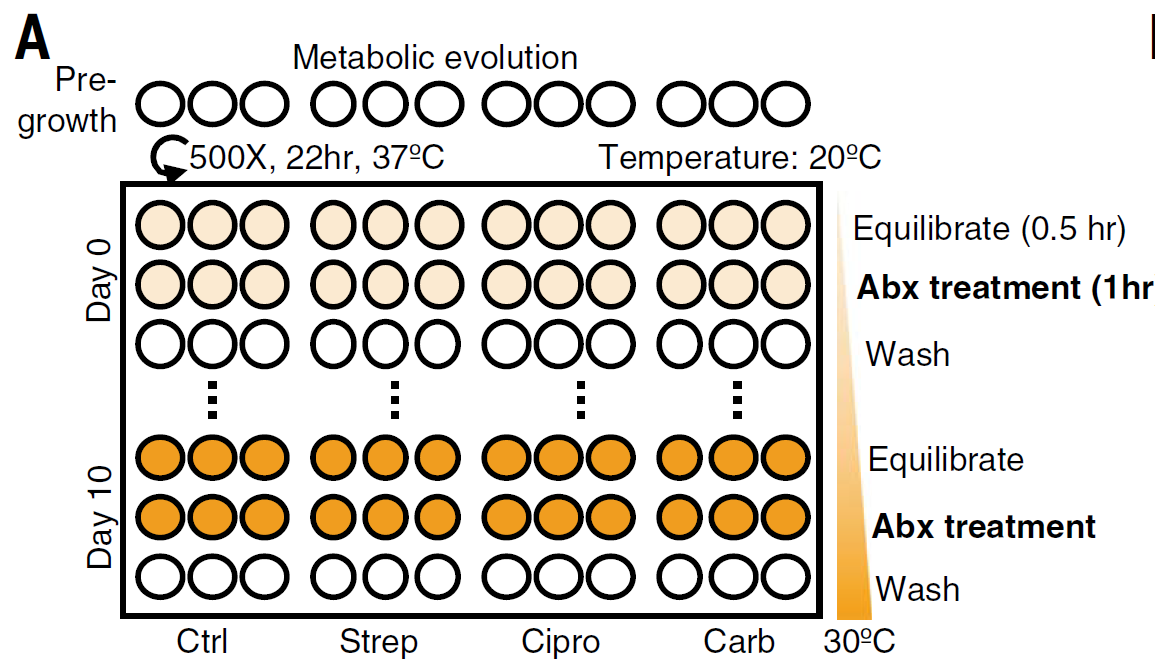
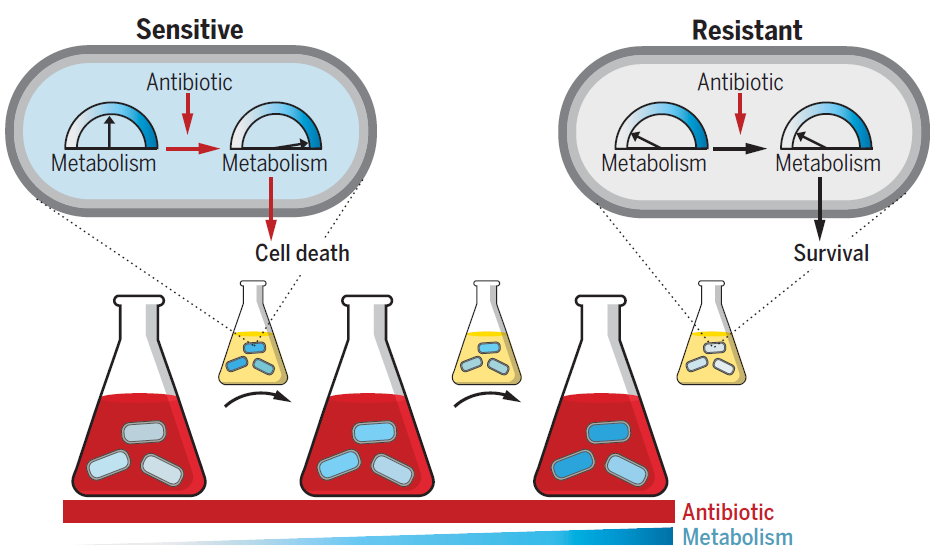
Figure 3: Altered metabolic state confers antibiotic resistance. Cells were exposed to high antibiotic concentrations (red) for short durations under incrementally increasing metabolic states (blue), separated by rounds of drug-free growth (small flasks). Left to right indicates evolutionary time. Initially, antibiotic-mediated metabolic stimulation partially contributes to cell lethality (sensitive cell). Evolved cells acquire resistance caused by decreased basal metabolic activity that avoids antibiotic-mediated stimulation and subsequent lethality (resistant cell).
Quick Biology can assist you with microbial NGS. Find More at Quick Biology.
See resource:
1. Lopatkin, A. J. et al. Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 371, (2021).



