Introduction
100-QuickPathTM PCR Arrays
Quickly Profiling Gene Expression in 100 Pathways for Gene Function Studies
PCR arrays are the most reliable tools for parallel quantitative analysis of gene expression signatures of a focused panel of genes. With Quick Biology's 100-QuickPathTM PCR Arrays, you can quickly analyze the expression of the most relevant genes in your pathway of interest with complete confidence in your results, to quickly gain a better biological insight in your study. The PCR Array system comprises a complete experimental workflow from sample preparation to data analysis and interpretation.
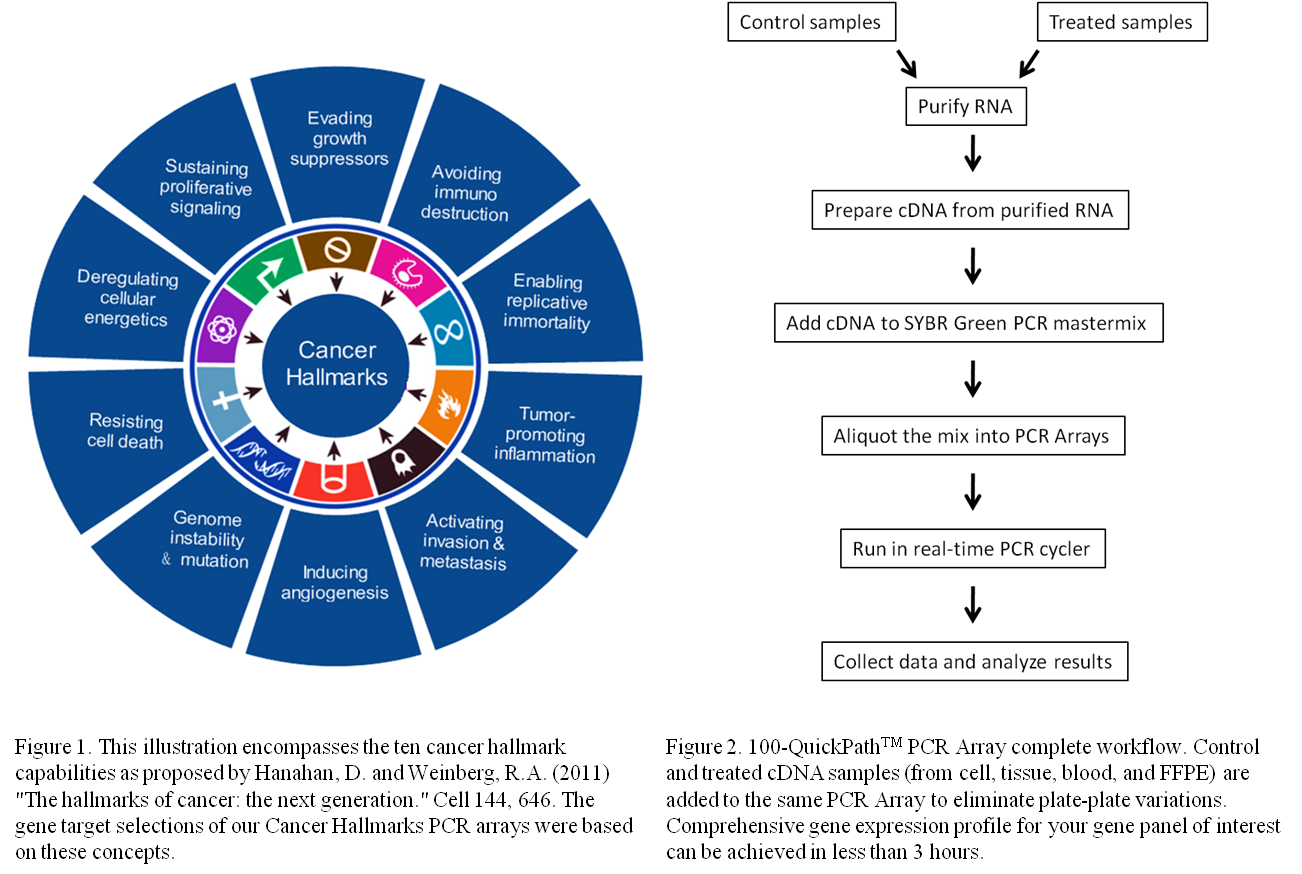
Quick Biology's first set of three PCR Arrays (Cancer Hallmarks) are designed to analyze genes involved in the hallmarks of cancer comprise 10 biological capabilities (Figure 1). In each hallmark, there are about 10 important pathways or biological processes. We designed highly specific primers based on these important transcription factor/oncogenes in these hallmark pathways to evaluate their expressions. You can quickly analyze the expression of the most relevant cancer hallmarks genes and to build a better biological story.
Order Information:
|
Product |
Genes in Each PCR Array |
Cat. #. |
Unit Price (USD) |
|
QV0301 |
$259.00 (Please specify your real-time PCR system plate specification when ordering)
|
||
|
QV0302 |
|||
|
QV0303 |
Please download the PCR Array manual and analysis template here:
![]() PCR Array Analysis worksheet_v3.xlsx
PCR Array Analysis worksheet_v3.xlsx
Please click the corresponding PCR array product to see the detailed plate design and gene targets.
Quick Biology's 100-QuickPathTM PCR Arrays provide:
l Quickly profiling gene panel of interest for any lab with a real-time PCR
l Compare gene expression of control and treated sample in the same array in less than 3 hours, eliminating plate-plate variation.
l Quickly analyze and publish your data using free easy-to-use data analysis template in excel format.
Why Use PCR Array?
l Quickly obtained biological insights by profiling a panel of genes expression related to a pathway or disease state.
l Simple and Accurate real-time PCR methods provide high sensitivity and wilder quantification ranges.
l Real-time PCR is fully quantitative and exhibit very good reproducibility.
l Easily applied to high throughput and routine use in any lab with a real-time PCR system.
How it works:
--Simple and easy workflow
As depicted in figure 2, simply mix your cDNA template with the appropriate ready-to-use PCR master mix, aliquot equal volumes to each well of the same plate, and then run the real-time PCR cycling program. Our PCR Arrays are made to be compatible with all common real-time PCR systems such as ABI, Bio-Rad, Eppendorf, QIAGEN, Roche, and Stratagene systems. Please find PCR array product manual files [ here ] ![]() PCR array manual_V1.2.pdf.
PCR array manual_V1.2.pdf.
--Ready-to-use data analysis
After you obtained Ct values from your real-time PCR system, simply Download the free Excel-based data analysis template [ here ] ![]() PCR Array Analysis worksheet_v3.xlsx.
PCR Array Analysis worksheet_v3.xlsx.
Copy your raw Ct values into the corresponding template cells. Data analysis is based on the ΔΔCt method with normalization of the raw data to either housekeeping genes on the PCR Array. The results will be transformed into figures including fold change (ranked) bar plots, and scatter plot, as shown below in the case studies.
Application use cases:
The PCR Arrays have increasingly used in many areas in life science and biomedical research such as cancer, immunology, stem cells, toxicology and miRNA research. Quick Biology's 100-QuickPathTM PCR Arrays are primarily focus in cancer research. The cancer hallmark PCR Array set are designed to analyze genes and pathways involved in the hallmarks of cancer comprise 10 biological capabilities. The following selected papers highlights the benefits of using PCR array for cancer research:
Wang R, Chadalavada K, Wilshire J, Kowalik U, Hovinga KE, Geber A, Fligelman B, Leversha M, Brennan C, Tabar V. Glioblastoma stem-like cells give rise to tumour endothelium. Nature. 2010 Dec 9;468(7325):829-33.
McElroy MK, Kaushal S, Tran Cao HS, Moossa AR, Talamini MA, Hoffman RM, Bouvet M. Upregulation of thrombospondin-1 and angiogenesis in an aggressive human pancreatic cancer cell line selected for high metastasis. Mol Cancer Ther. 2009 Jul;8(7):1779-86. Epub 2009 Jul 7.
Bose RN, Maurmann L, Mishur RJ, Yasui L, Gupta S, Grayburn WS, Hofstetter H, Milton T. Non-DNA-binding platinum anticancer agents: Cytotoxic activities of platinum-phosphato complexes towards human ovarian cancer cells. Proc Natl Acad Sci U S A. 2008 Nov 25;105(47):18314-9. Epub 2008 Nov 19
Chen N, Ye XC, Chu K, Navone NM, Sage EH, Yu-Lee LY, Logothetis CJ, Lin SH. A secreted isoform of ErbB3 promotes osteonectin expression in bone and enhances the invasiveness of prostate cancer cells. Cancer Res. 2007 Jul 15;67(14):6544-8 DiMeo TA, Anderson K, Phadke P, Fan C, Perou CM, Naber S, Kuperwasser C. A novel lung metastasis signature links Wnt signaling with cancer cell. Cancer Res. 2009 Jul 1;69(13):5364-73. Epub 2009 Jun 23.
Santisteban M, Reiman JM, Asiedu MK, Behrens MD, Nassar A, Kalli KR, Haluska P, Ingle JN, Hartmann LC, Manjili MH, Radisky DC, Ferrone S, Knutson KL. Immune-induced epithelial to mesenchymal transition in vivo generates breast cancer stem cells. Cancer Res. 2009 Apr 1;69(7):2887-95. Epub 2009 Mar 10.



