Introduction
PCR Array Plate layout:
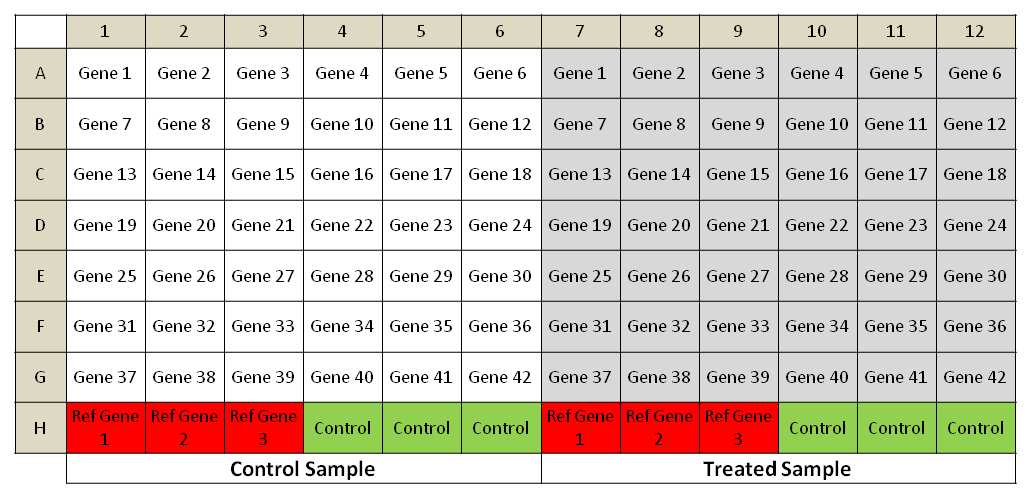
PCR Array plates design for Ten cancer hallmarks and target genes:
| Array # | Cancer Hallmark Pathways | Census Cancer Genes |
| Array #1 | Sustaining Proliferative Signaling | |
| --MAP kinase pathway | MAP2K1, MAP2K2 | |
| --PI3K/AKT signaling | AKT1, AKT2, MTCP1, PIK3CA, PIK3R1, PTEN, TCL1A | |
| --RAS signaling | JUN, FOS | |
| --PTEN signaling | AKT1, CDKN1B, FOXO3, PTEN | |
| mTOR pathways | AKT1, PTEN, TSC1, TSC2 | |
| --Evading Growth Suppressors | ||
| p53 Signaling Pathway | ||
| --p53 Activation | TP53, ATM, ATR, BRCA1, CDKN2A, CHEK2, FOXO3, MDM2, MSH2 | |
| --p53 Targets for apotosis | EGFR, ESR1, FAS, TP53 | |
| RB1 Signaling Pathway | ||
| --RB1 Activation | CCNE1, CDK4, CDK4, CCND2, CCND3, CCND1 | |
| --RB1 downstream targets for cell cycle arrest | RB1, MDM2 | |
| TGFB signaling pathway | EP300 | |
| LKB1 signaling pathways | STK11 | |
| NF2 Contact Inhibition | NF2, TAZ, YAP1 | |
| Resisting Cell Death | ||
| --Pro-Apoptotic | ABL1, CYLD, FAS, TP53 | |
| --Anti-Apoptotic | AKT1, BCL2, BIRC3 | |
| --Death Domain Receptors | CRADD, DAPK1, FADD, TRADD | |
| Array #2 | --Autophagy | AKT1, BCL2, ESR1, FAS, TP53 |
| --Necrosis | CYLD | |
| --Caspases | CASP8 | |
| --Caspase Activation | TP53 | |
| --Caspase Inhibition | CD27, XIAP | |
| Enabling Replicative Immortality | ||
| --Senescence Pathway | ATM, CCND1, CCNE1, CDK4, CDK6, CDKN2A, CHEK2, MDM2, RB1, TP53 | |
| --p53 / pRb Signaling & the Cell Cycle | ABl1, AKT1, CDKN2C, MYC, PIK3CA | |
| --Telomere Maintenance | BLM, ERCC4, MYC, NBN, POT1, TERT | |
| --Telomerase Regulation | ABL1, AKT1, BCL2, KRAS, MEN1, MYC, PAX8, PPARG, PPP2R1A, RB1, TP53 | |
| --Telomere-Associated Complexes | POT1, TERF2, TERT, ERCC4, MSH2, TERF2 | |
| --Other Telomere-Associated Genes | ATM, CHEK2, HNRNPA2B1, HSP90AA1 | |
| Inducing Angiogenesis | ||
| --Growth Factors & Receptors | KDR | |
| --AKT & PI3 Kinase Signaling | AKT1, AKT2, PIK3CA, PIK3R1 | |
| --Small G-Protein Signaling | HRAS, KRAS, NRAS, RAC1, RAF1 | |
| --Inhibition of Angiogenesis by TSP1 | JUN | |
| --MAP Kinase Signaling | MAP2K1, MAP2K2 | |
| --NFAT Signaling | NFATC2 | |
| --Angiogenesis Transcription Factors | ARNT, NFATC2 | |
| Activating Invasion and Metastasis | ||
| --JAK-STAT signaling | JAK1, JAK3, STAT3, STAT5B, STAT6, PTPN11, PTPRC, GATA2, SOCS1, SMAD4, FAS | |
| --MAPK pathway | MAP2K1, MAP2K2 | |
| --Wnt signaling pathway | APC, AXIN1, AXIN2, BCL9, CTNNB1, WIF1, AXIN2, CCND1, CCND2, JUN, MYC | |
| Array #3 | --Cell Adhesion Molecules | APC, CDH1, CDH11, ITGA7, SYK, ITGA7 |
| Extracellular Matrix Molecules | ||
| --Regulation of the Cell Cycle | APC, CDKN2A, HRAS, KRAS, NF2, PTEN, RB1, TP53 | |
| --Negative Regulation of Cell Proliferation | CDKN2A, MDM2, NF2 | |
| --Positive Regulation of Cell Proliferation | TSHR | |
| --Transcription Factors & Regulators | ETV4, EWSR1, MYC, MYCL, NR4A3, RB1, SMAD4, TP53 | |
| Deregulating cellular energetics | ||
| --TCA Cycle | FH, IDH1, IDH2, SDHB, SDHC, SDHD | |
| --HIF1 & Co-Transcription Factors | ARNT, NCOA1, PER1, TP53 | |
| --MYC & Co-Transcription Factors | MAX, MYCL, MYCN | |
| Avoiding immune destruction | ||
| --Innate Immunity | HLA-A, MYD88 | |
| --Th2 Markers & Immune Response | GATA3, STAT6 | |
| --Th17 Markers: | STAT3 | |
| --Cytokines | IL2 | |
| --Other Adaptive Immunity Genes: | HLA-A, JAK2 | |
| --Defense Response to Bacteria | MYD88 | |
| --Defense Response to Viruses | HLA-A | |
| Tumor promoting inflammation | ||
| --Inflammatory Response | STAT3 | |
| --Pro-Inflammatory Genes | IL2 | |
| --Antigen Presentation | HLA-A | |
| --Interferon-Responsive Genes | MYD88 | |
| --Chemokine Receptors | ACKR3 | |
| --Interleukins | IL2 | |
| --STAT Targets | MYC | |
| Genome instability & mutation | ||
| --ATM / ATR Signaling | ATM, ATR, BRCA1, CHEK2, FANCD2, TP53 | |
| --Nucleotide Excision Repair | DDB2, ERCC2, TP53, XPA, XPC | |
| --Base Excision Repair | TP53 | |
| --Mismatch Repair | ABL1, MLH1, MSH2, PMS1, PMS2 | |
| --Double-Strand Break Repair | ATM, BLM, BRCA1, MLH1, NBN | |
| --Other DNA Repair Genes | ATR, ATRX, BRIP1, CHEK2, FANCA, FANCD2, FANCG, RAD21, RAD51B |



