2020-11-2 By Quick Biology Inc.
Next-generation sequencing (NGS) has changed diagnostic workflows and provided an unprecedented, simple way of discovering rare genomic diseases (QB News). Through whole genome sequencing (WGS) or whole exome sequencing (WES), scientists can trace genetic defects to mutations in a single gene. However, as only ~ 1% of the human genome is the coding region, many polymorphisms discovered from patients remain undiagnosed, and such genomic screening diagnostic rates are only 25-30%.
In The Journal of Clinical Investigation, scientists incorporated transcriptome sequencing in the genetic diagnostic toolbox (Fig.1, ref1). This design bases on the advantages of transcriptomes, which includes:
- 1. RNA is an intermediate between genetic material DNA and phenotype proteins.
- 2. RNA changes can be caused by variants occurred in non-coding regions, nonsynonymous site, or intergenic regions.
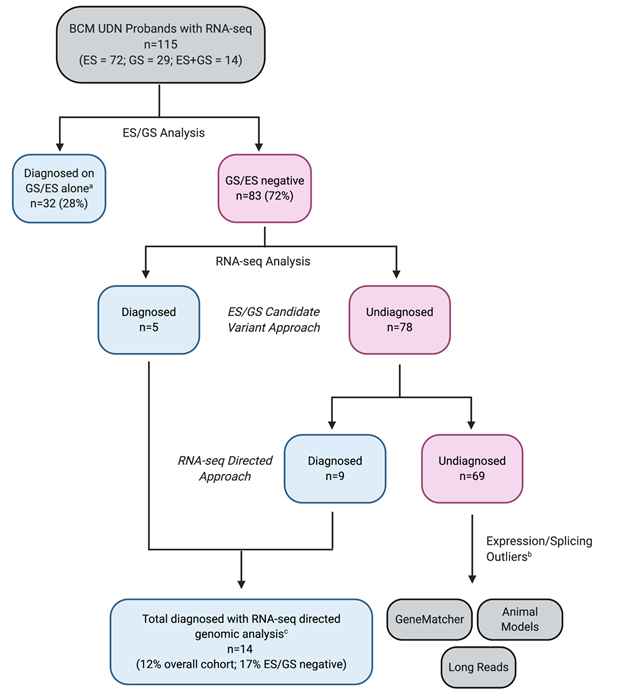
Both things aid in the prioritization and resolution of genetic mutations of uncertainty. A traditional analytic strategy relies on the identification of mutations using exome or whole genome data first, then reviewing transcriptome to determine any functional consequences (i.e. transcriptome change) of certain variants. Here, the authors described a new approach using RNA-seq first to direct downstream genomic analysis and diagnose patients with rare, Mendelian conditions. Comparing to the traditional approach (i.e. candidate-driven approach), RNA-seq direct approach improves diagnostic rate, particularly in ES (exon sequencing)/CMA (chromosomal microarray)-negative patient cases (Fig.2).
Figure 1: Transcriptome-directed Genomic Analysis
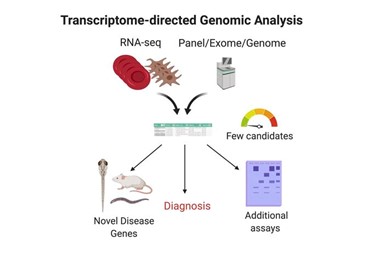
Figure 2: Flow diagram outlining BCM (Baylor College of Medicine) UDN clinical site RNA-seq diagnostic research process.
- a. Cases diagnosed on an initial review of ES/GS without needing RNA-seq.
- b. Undiagnosed but with expression/splicing outliers prompting follow-up studies for potential novel disease gene discovery.
- c. Five cases diagnosed with ES/GS candidate variant approach were validated using RNA-seq directed approach. (ES: exome sequencing, GS: genome sequencing).
Are you ready to sequence your samples (whole genome, whole exome, or transcriptome) of interest? Find More at Quick Biology.
Reference:
- 1. Murdock, D. R., Dai, H., Burrage, L. C., Rosenfeld, J. A., Ketkar, S., Müller, M. F., … Lee, B. (2020). Transcriptome-directed analysis for Mendelian disease diagnosis overcomes limitations of conventional genomic testing. Journal of Clinical Investigation. https://doi.org/10.1172/jci141500



