2019-11-17
The nervous system exhibits the most extensive usage of pre-mRNA alternative transcript isoforms. However, it remains debated as to what extent pre-mRNA alternative splicing events detected by RNA-seq are indeed recruited for translation to produce proteins. Ribosome-seq uses specialized messenger RNA sequencing to determine which mRNAs are being actively translated (Fig. 1), it only targets mRNA protected by the ribosome during the process of decoding by translation.
In recent Nature Neuroscience, Researchers in Switzerland conducted large-scale tagged-ribosomal affinity purification (RiboTRAP) of ribosome-associated mRNAs from genetically defined neurons of the mouse forebrain (Fig. 2a, b, ref1). Their dataset provides an extensive resource of transcript diversities across major neuron classes (Fig.3). Their work also revealed that alternative splicing was independently regulated other than its own gene level, playing important roles in synaptic interactions and neuronal architecture (Fig. 4).
Figure 1: Ribosome Profiling in brief. (https://en.wikipedia.org/wiki/Ribosome_profiling)
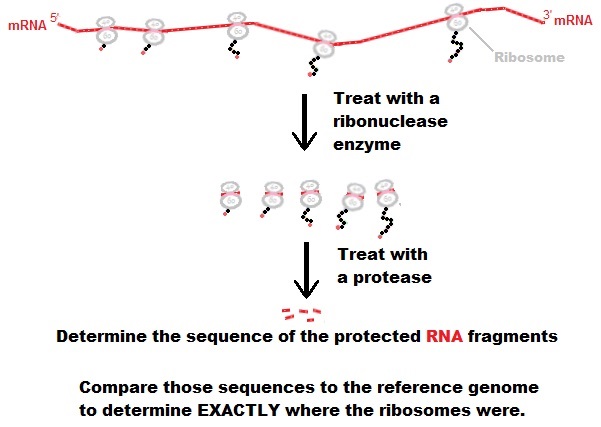
Figure 2: (a) Schematic representation of Ribo TRAP pull down; (b) cartoons representing neuronal cell populations isolated from mouse neocortex and hippocampal samples). (ref1)
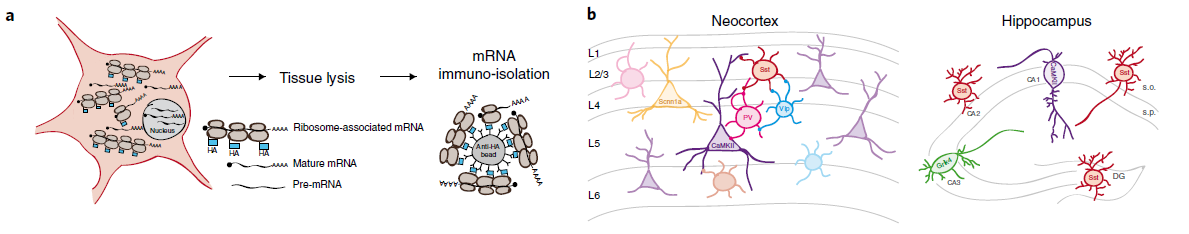
Figure 3: Alternative splicing and alternative TSS usage drive transcript diversification in neocortical neurons. (a) Histogram representing different types of alternative splicing. (b) examples of alternative splicing and alternative TSS.
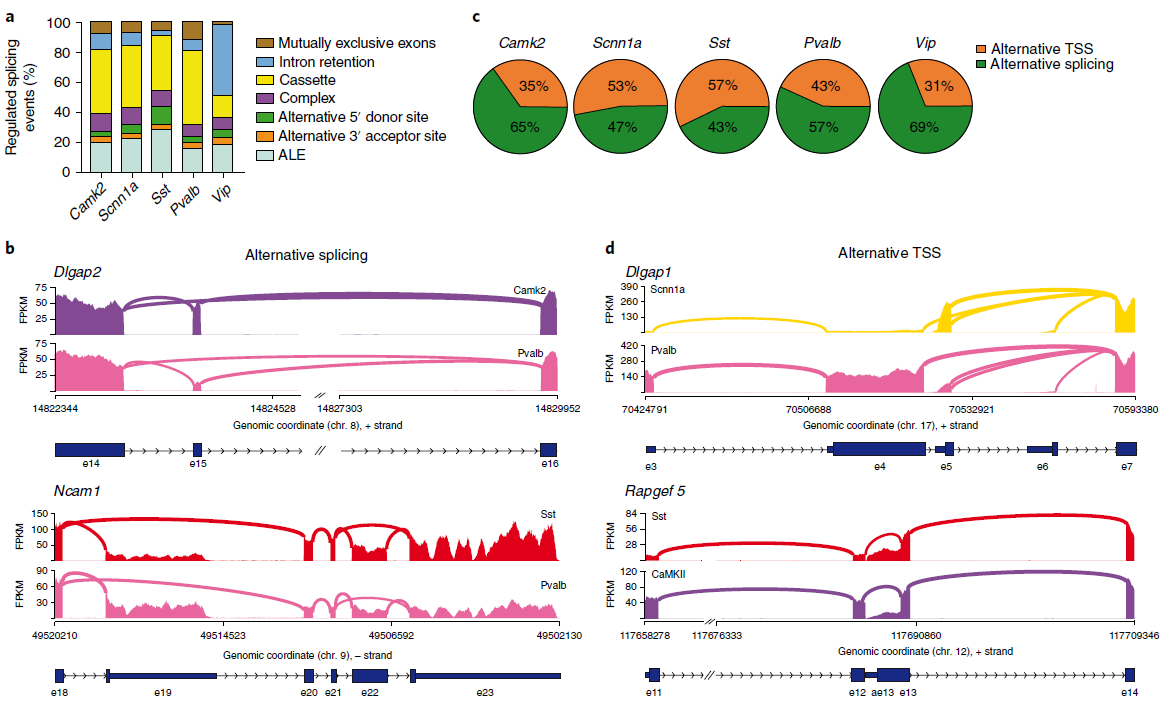
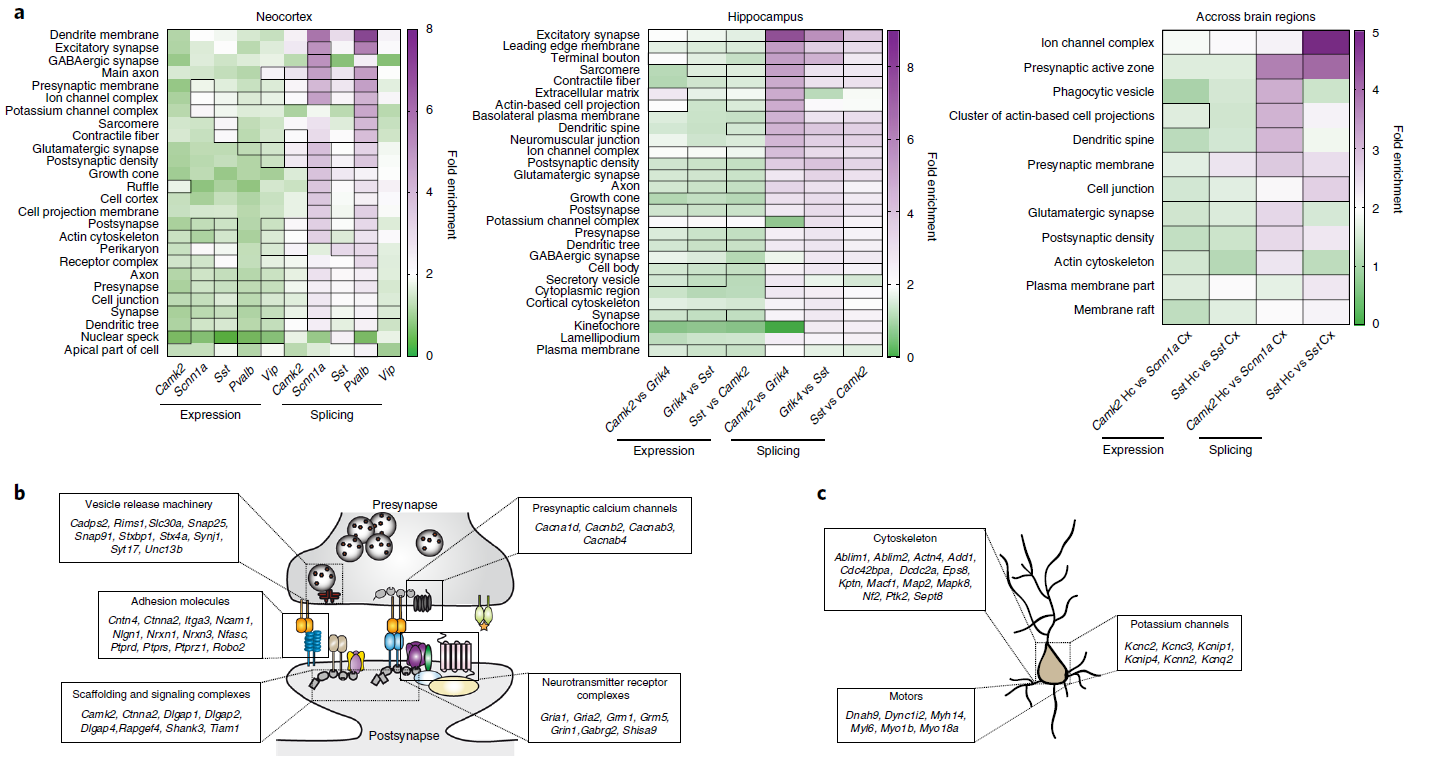
Figure 4: Alternative splicing programs are highly dedicated to the control of synaptic interactions and neuronal architecture. (a) Heatmap representing the fold-enrichment of GO terms for transcripts at gene level or alternative splicing level. (b) Cartoon of main categories of genes or alternative splicing.
Quick Biology is an expert in using NGS to identify alternative splicing. Find More at Quick Biology.
Reference:
- 1. Furlanis, E., Traunmüller, L., Fucile, G. & Scheiffele, P. Landscape of ribosome-engaged transcript isoforms reveals extensive neuronal-cell-class-specific alternative splicing programs. Nat. Neurosci. 22, 1709–1717 (2019).



