2019-08-18 by Quick Biology Inc.
Deconstruction of gene regulation is critical for cell development and human diseases. Cell type-specific gene expression in eukaryotic cells is regulated not only cis-acting DNA elements (such as promoter, mRNA splicing sites), chromatin structure (accessible for transcription factors’ binding) is another level controlling gene expression.
Chang Group in Stanford University developed the assay for transposase-accessible chromatin using sequencing (ATAC-seq) in 2013, an advanced method for MNase-seq, FAIRE-seq, and DNAse-seq (1,2). This technique is based on the fact that Tn5 transposase can excise any open regions of the genome. By inserting sequencing adapters in the cut site, the tagged DNA fragments can be amplified and sequenced (Fig.1) (2).
Drop-seq is the single-cell RNA sequencing method, which uses a microfluidic device to compartmentalize droplets containing a single cell with other library preparation reagents (3).
In recent Nature Biotechnology, Chang group adopts droplet technique (Fig.2), perform single-cell ATAC-seq in human immune cell development and intratumoral T cell exhaustion (4). By analysis scATAC-seq profiles, researchers identify chromatin regulators of therapy-responsive T cell subsets, reveal a shared regulatory program that governs intratumoral CD8+ T cell exhaustion and CD4+ T follicular helper cell development (Fig. 3).
Figure 1: ATAC-seq workflow (2)
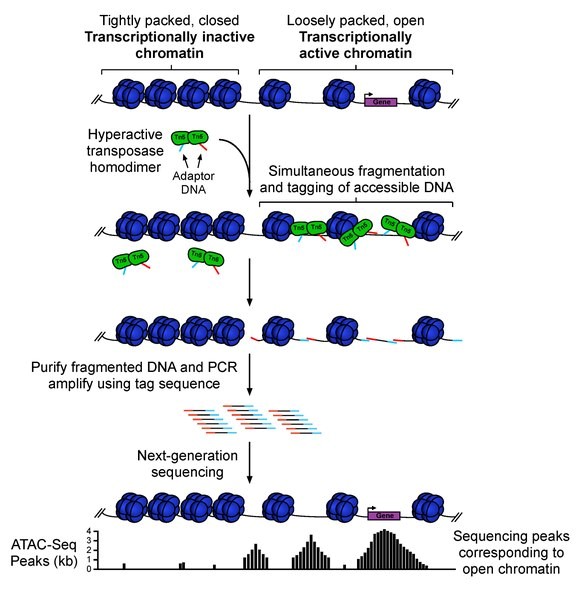
Figure 2: Drop-seq Microfluidic procedure (4).
Figure 3: Schematic of regulatory modules controlling TEx and Tfh differentiation (4).
Quick Biology has been providing ATAC-seq services for over 3 years. We have great ATAC-seq experience with low cell number and low mitochondrial DNA contamination. Are you ready to sequence your samples of interest? Find More at Quick Biology.
Reference:
- 1. Buenrostro, Jason D; Giresi, Paul G; Zaba, Lisa C; Chang, Howard Y; Greenleaf, William J (2013). "Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position". Nature Methods. 10 (12): 1213–1218
- 2. Wikipedia about ATAC-seq: https://en.wikipedia.org/wiki/ATAC-seq#cite_note-BuenrostroGiresi2013-1
- 3. Drop-Seq: Macosko E. Z., Basu A., Satija R., Nemesh J., Shekhar K., et al. (2015) Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161: 1202-1214
- 4. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. 1. Nat Biotechnol. 2019 Aug;37(8):925-936.



