Introduction
Micro RNAs (miRNAs) are a class of small, endogenous RNAs of 21–25 nucleotides (nts) in length, non-coding RNAs that mediate protein expression through transcript degradation, inhibition of translation, or sequestering transcripts. Alterations in miRNA can have many effects on gene expression changes in development, differentiation, signal transduction, infection, aging, and disease. Up to date, many research studies have shown that circulating miRNA expression associated with both normal and disease biology as miRNAs expressed in virtually all biofluids, including serum, plasma, cerebrospinal fluid (CSF) and urine. Specifically, with cancers, numerous studies and reviews have identified the presence of various miRNAs affecting cancer cell proliferation, resistance to apoptosis, invasiveness, and differentiation.
Quick Biology’s microRNA sequencing (miRNA-seq) service enables the reliable investigation of the micro RNAs from a normal to a low amount of miRNA with minimal bias. Due to the growth of circulating miRNAs as potential biomarkers, Quick Biology’s microRNA seq pipeline is optimized to map miRNA down to ultralow input levels. In addition, we integrate Unique Molecular Indices (UMIs) into the reverse-transcription process, enabling unbiased and accurate miRNome-wide quantification of mature miRNAs by NGS. Collectively, Quick Biology’s miRNA offers an unrivaled Sample to Insight solution for differential expression analysis and discovery of novel miRNAs using next-generation sequencing.
Request a miRNA-Seq Quote, Click Here!
Quick Biology’s miRNA-seq service delivers:
· Comprehensive discovery and differential expression of miRNA from many sample types including biofluids, low input samples, FFPE and any tissue.
· Gel-free miRNA sequencing library prep from as little as 1 ng of total RNA
· Integrated Unique Molecular Indices (UMIs) enable quantification of individual miRNA molecules, eliminating PCR and sequencing bias
· miRNA-seq data includes both miRNAs and other small RNAs (piRNA)
· Bioinformatics solutions for differential expression analysis and discovery of novel miRNAs
Quick Biology’s miRNA-seq is suitable for small RNAs for the analysis of:
(1) Biofluids (such as serum, plasma, CSF and urine)
(2) Total RNA and miRNA
(3) Cells, fresh/frozen tissue and FFPE tissues
(4) Exosomes containing small RNAs
Quick Biology miRNA-seq workflow:
We follow a complete optimized workflow from cell lysis, miRNA isolation, and cDNA synthesis with Unique Molecular Index (UMI) – with negligible sequence bias.
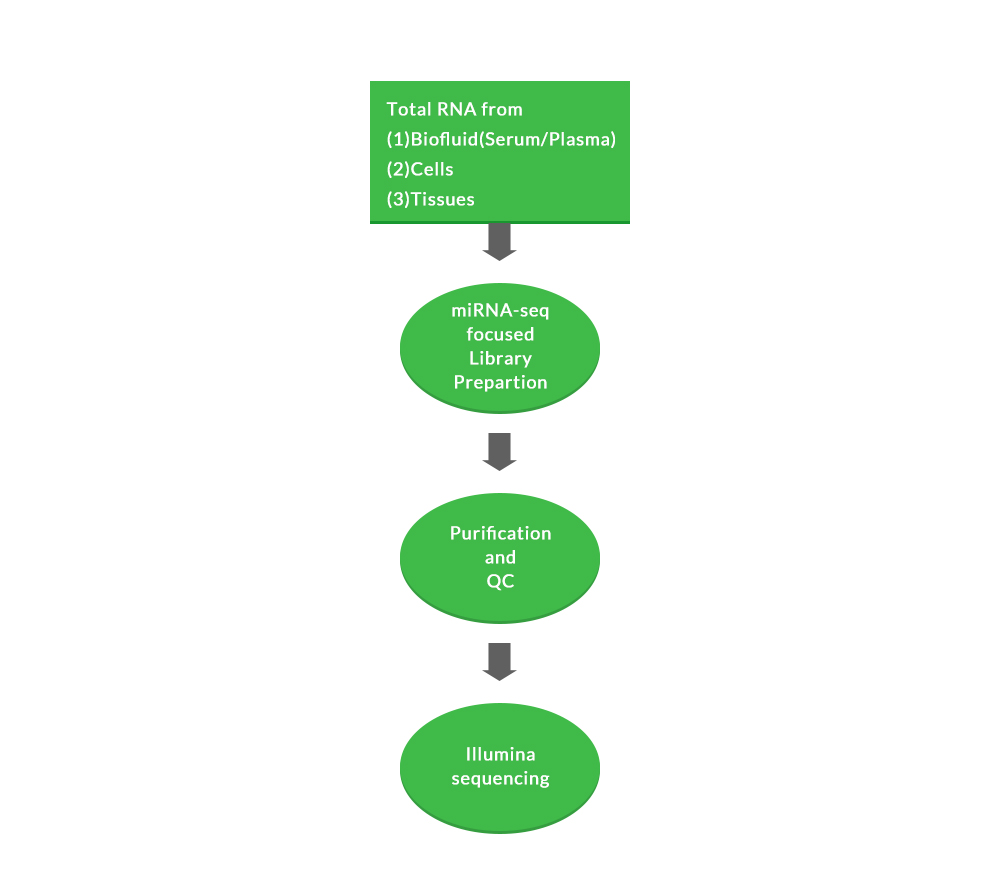
Sample requirements:
· Serum, plasma, cells, fresh/frozen tissue and FFPE tissues
· 100 ng – 500 ng (minimum 1 ng) of purified total RNA (please isolate RNA using kits that retain miRNA fractions)
Bioinformatics for miRNA Sequencing:
Standard Analysis:
a. Raw data QC and clean up
b. Alignment to a reference with mapping statistics
c. miRNA count, quantifications, and differential miRNA expression analysis
d. Downstream miRNA target analysis and clustering
f. Final project report (HTML) with analysis methods, publication-ready graphics, and references
Advanced Analysis:
Please inquire about Quick Biology's customized data analysis.
Frequently Asked Questions:
1. Total RNA from what sample types are compatible with Quick Biology’s miRNA-seq?
Total RNA from cells, fresh/frozen tissue, FFPE tissue, serum/plasma, blood and other fluids are compatible with Quick Biology’s miRNA-seq. We can provide sample isolation service.
2. Should total RNA or small enriched RNA be used as the starting material for the Quick Biology’s miRNA-seq?
Total RNA containing miRNA is the required starting material for the Quick Biology’s miRNA-seq. It is not necessary to enrich for small RNA.
3. What are the concerns of adapter dimers in miRNA-seq library constructions?
One of the biggest limitations to the small RNA library protocol are the insert-less 5’adapter and 3’adapter ligations that form what are called adapter dimers. These dimers compete for resources during RT-PCR. When over-amplified, these ~120 bp products can contaminate true 150 bp microRNA library bands. Quick Biology’s optimized protocol utilizing modified oligonucleotides is used to eliminate the reverse-transcription of adapter dimers.
4. Should I pool or multiplex samples for small RNA sequencing?
Yes! We can multiplex up to 48 miRNA libraries in one run of Illumina sequencing to save time and cost. To determine the level of multiplexing or number of libraries to pool together, you should consider whether you’re interested in differential expression, microRNA profiling or discovery of novel/new microRNAs. The latter application, discovery, will require reads between 5-10 million, while profiling studies typically only require 100K – 1M reads per library.
5. Does sequencing microRNA and mRNA require separate library construction protocols?
Yes! when studying differential expression of both mRNA and small RNAs, two separate library constructions (protocols) must be performed on the same total RNA sample. Providing total RNAs, we can isolate both miRNA and mRNA from your samples.
6. What sequencing read length is recommended for microRNA sequencing?
MicroRNAs are short ~20-30 base RNA species, we recommend to sequence small RNA libraries using a 1x50 or 1x75 bp sequencing run.
7. How many reads are required for small RNA sequencing?
For miRNA expression profiling, 100K–5M mapped reads per sample is typically an accepted range. For miRNA discovery applications, 10-20M reads should be considered.
Request a miRNA-Seq Quote, Click Here!
Turnaround Time
About 4 weeks, depends on the quality of submitted samples, one additional week for data analysis.



