2020-07-13 by Quick Biology Inc.
The COVID-19 pandemic has already greatly disrupted all our life and society since the outbreak about seven months ago. As of July 13th, SARs-CoV2 has already resulted in over 3.4 million confirmed cases and 137,786 deaths in the USA (https://coronavirus.jhu.edu/map.html). Something worse is happening, at the beginning of June 2020, as more and more states are moving to reopen their economies. Such ease of coronavirus restrictions planning for the economic and society reopening, many of the states, especially states especially Florida, Arizona, and Alabama are experiencing a surge in cases. To adequately combat the COVID-19 pandemic and reopen society safely, comprehensive, and reliable SARs-CoV2 screening is essential to guide the community to contain the spread.
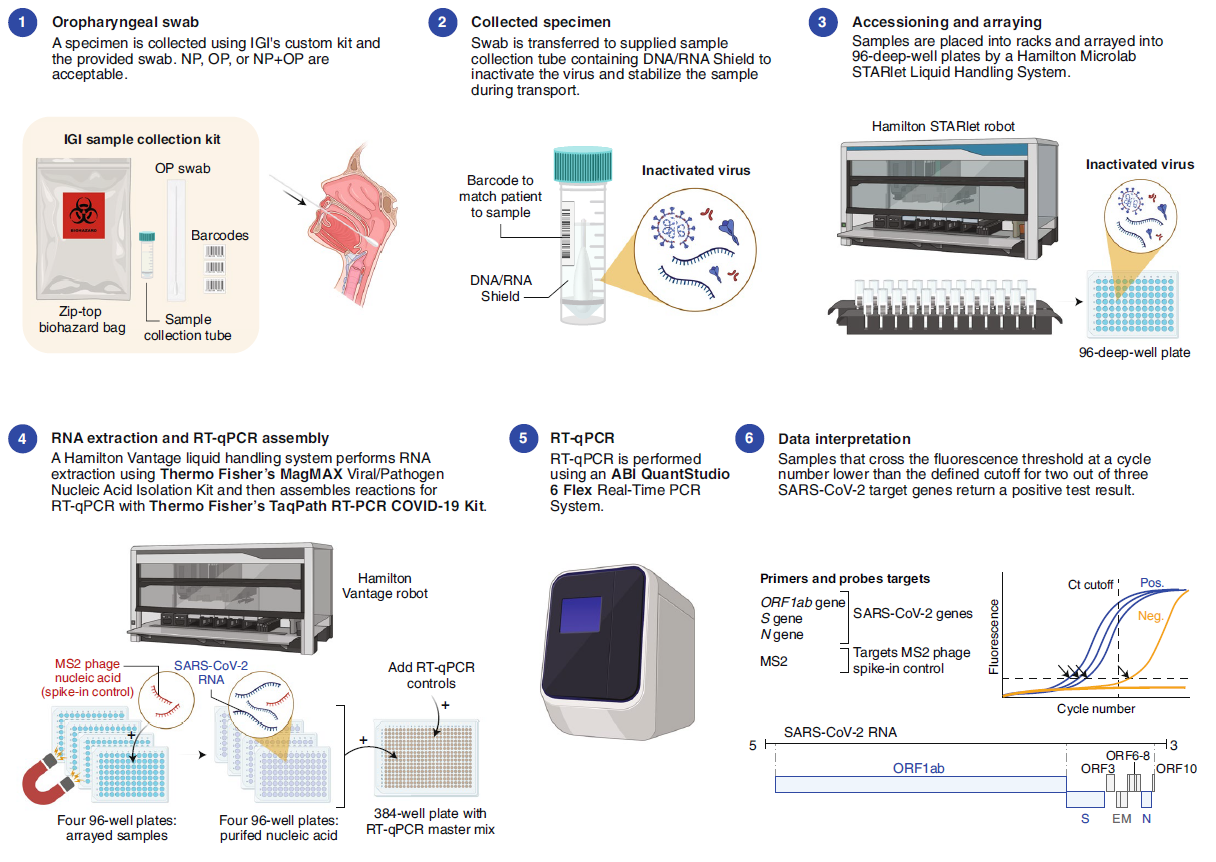
On June 18, Nature Biotechnology posted two Correspondence letters about the road map of SARs-CoV2 detection (ref1, 2). One shows how a European research Institute (Crick COVID-19 Consortium---- CCC) in collaboration with partner hospitals and diagnostic laboratory, builds up a testing pipeline from virus handling, robotics, molecular testing to final informatics/report to patients (Fig.1). The key reagent CCC used is from Shenzhen-headquartered BGI, a Chinese genome sequencing company. The CCC pipeline (Fig. 1) is capable of 2,500-3,000 samples in approximately 24 hours. The other shows how an American research Institute (the Innovative Genomics Institute (IGI) at UC Berkeley) established a clinical test for SARs-CoV-2 in three weeks (Fig.2). The key reagent IGI used is from Thermo Fisher. To lower the cost of the test, IGI researcher did a half reaction volume, modified sample collection kit with the goals of circumventing supply chain shortages (Fig.2), the IGI test capacity is 1,000 tests per day.
Figure 1: Schematic of the CCC test and reporting pipelines. EDS, experiment document single; sFTP, secure file transfer protocol (ref1). a, Specimen barcodes are scanned at sample reception, before viral inactivation in a class I or II safety cabinet, processing through RNA extraction using an in-house protocol and RT-PCR testing using a commercial kit (BGI). The number of samples processed through the pipeline per day has ranged from 39 (1 plate) to 1,386 (15 plates). b, CCC reporting pipeline. Test results are reported continuously through a custom-made remote web application, allowing remote clinical scientists and pathologists working outside the institute to authorize reports, in line with the established SOP. EDS, experiment document single; sFTP, secure file transfer protocol.

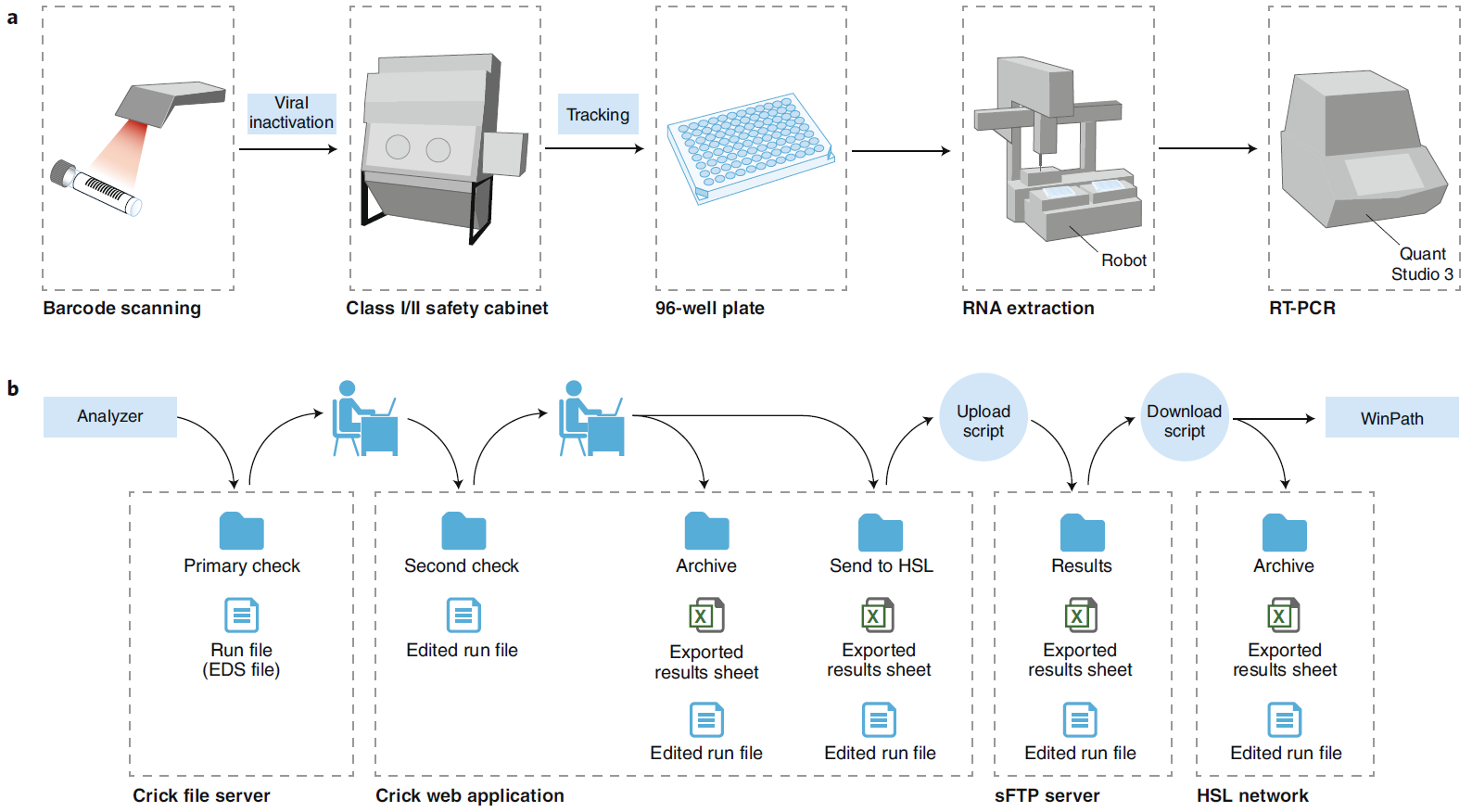
Figure 2: Overview of the IGI SARS-CoV-2 Testing Consortium assay (ref2).
Both platforms/pipelines mentioned above are based on RNA extractions and laboratory real-time quantitative-PCR. Quick Biology has been developing an extraction-free, laboratory-instrument-free SARs-CoV2 testing (Now in pre-EUA submission). Find More at Quick Biology.
Ref:
1. Aitken, J. et al. Scalable and robust SARS-CoV-2 testing in an academic center. Nat. Biotechnol. (2020). doi:10.1038/s41587-020-0588-y, (https://www.nature.com/articles/s41587-020-0588-y)
2. Pitts, P. J. Regulatory centaurs. Nat. Biotechnol. 38, 791–797 (2020). (https://www.nature.com/articles/s41587-020-0583-3)



