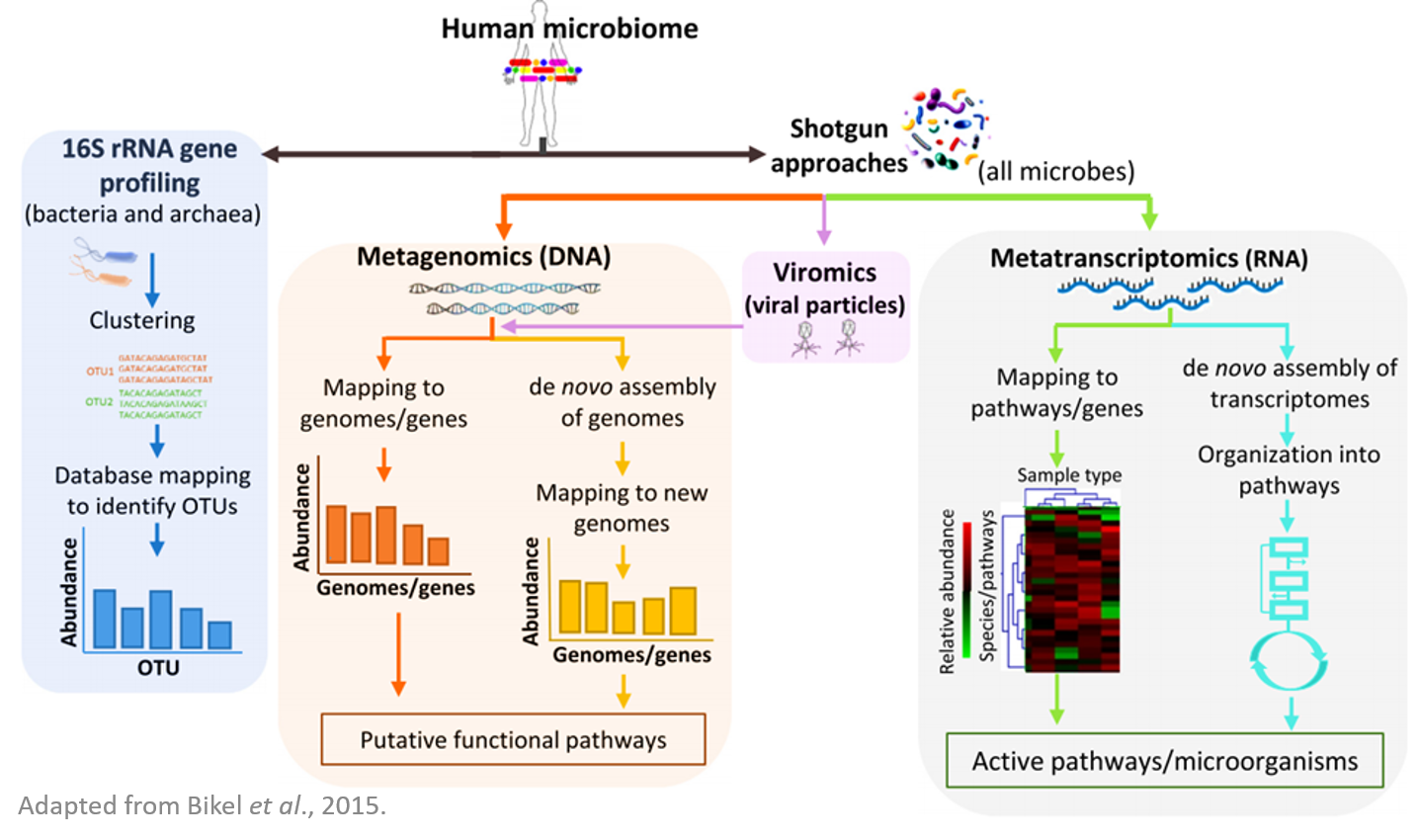
Metagenomics is the study of genetic material recovered directly from environmental samples. Metagenomics tools enable the population analysis of un-culturable or previously unknown microbes. The advance in high-throughput technologies used to analyze the components of the microbiome have substantially improved our knowledge of the microbial communities. For studying the microbial community of the microbiome using high throughput sequencing technologies, there are several types of large scale analyses: the 16S profiling analysis which is based in sequencing the hypervariable regions of the 16S rRNA gene and the shotgun analysis which is based in direct sequencing of the total DNA (metagenome) and/or total RNA (metatranscriptome). In addition, the viral component of the microbiome (virome) can be also analyzed by sequencing the total viral particles.
Quick Biology is offering end-to-end services using Illumina and third-generation sequencing platforms (PacBio and Nanopore) for microbial and viral metagenomic/metatranscriptomic sequencing projects. We provide 16S rRNA gene sequencing to identify and quantify bacterial taxa present within a microbiome; In the metagenomic sequencing analysis, the DNA sequences obtained can either be mapped to reference genomes/genes or used for de novo assembly of genomes. In viromics sequencing analysis, we sequence the virus genomes to analyze the active genes and species of the microbiome. We apply the metatranscriptomic analysis to identify the active pathways, genes, and microorganisms.
Request a metagenomics/metatranscriptomic sequencing Quote, Click Here!
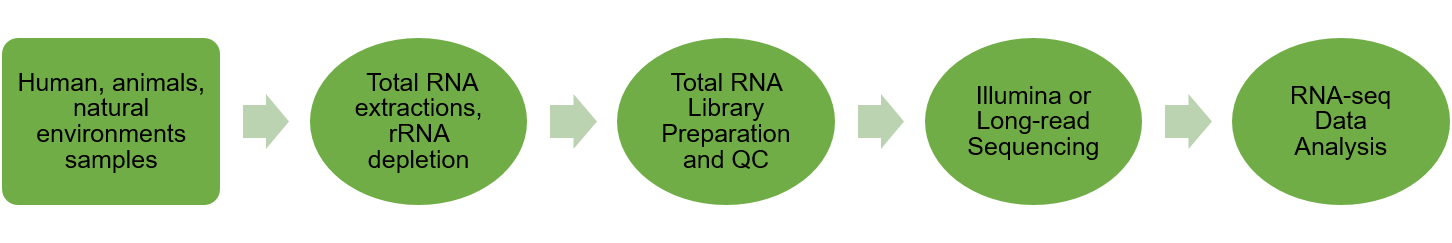
Quick Biology's Microbial and Virus Metagenomics Sequencing Workflow:
16S/18S/ITS Amplicon Sequencing
Our workflow not only provides you the flexibility of targeting any regions of 16S V1-V9 and ITS, but also increases coverage with novel primers to increase phylogenetic coverage of Bacteria and Archaea and enable species-level resolution for human microbiome profiling.

Metagenomic Shotgun Sequencing and Microbial Whole Genome Sequencing
Shotgun sequencing and WGS allows for the detection of low abundance members of microbial communities and are used to examine thousands of organisms in parallel and comprehensively sample all genes, providing insight into community biodiversity and function.

Metatranscriptomic sequencing
Our metatranscriptomic sequencing provides complete profiling of gene expression of microbes within natural environments, i.e., the metatranscriptome. It also allows us to obtain whole gene expression profiling of complex microbial communities.
Sample Requirements:
- - Cultured bacteria, fungi, phage or virus, and other microorganisms
- - Samples sources including human, animals, natural environments, and industrial environments
- - DNA or PCR products (10 ng - 100 ng; OD260/280=1.8~2.0; free of enzymatic inhibitors)
Sequencing Strategy:
- - Illumina Platform: Hiseq, PE150 for metagenomics and MiSeq, PE300/PE250, for 16s rRNA gene sequencing
- - PacBio and Nanopore long-read platform for full-length 16S/ITS rRNA or RNA sequencing and whole genome DNA sequencing
Quick Biology's comprehensive Microbial and Viral Sequencing Data Analysis:
- - 16S rRNA gene profiling, OTU annotations, clustering, alpha and beta diversity
- - De novo reference-based genome assembly, contigs generation, gene prediction, functional annotation and SNP/InDel identification
- - Viral genome identification from the host, taxonomy identification, and phylogenetic analysis
- - De novo or reference-based transcriptome assembly, gene annotation, differential gene expression analysis, and structure identifications.
- - More customized data mining upon your request based on your research interest.
Request a metagenomics/metatranscriptomic sequencing Quote, Click Here!

