CUT&RUN (cleavage under targets and release using nuclease) is an epigenomic profiling method and is performed in situ on immobilized, intact cells without crosslinking. DNA fragmentation is achieved using micrococcal nuclease fused to Protein A and/or Protein G (pA/G-MNase). The fusion protein is directed to the desired target through binding of the Protein A/G moiety to the Fc region of an antibody bound to the target. DNA under the target is subsequently cleaved and released and the pA/G-MNase-antibody-chromatin complex is free to diffuse out of the cell. DNA cleavage products are extracted and then processed by next generation sequencing (NGS). Quick Biology is now offering CUT&RUN sequencing (CUT&RUN-seq) with a proprietary immunotethering approach based on Chromatin ImmunoCleavage (ChIC) and Cleavage Under Targets and Release Using Nuclease (CUT&RUN) methods. This approach can be used to map histone post-translational modifications (PTMs) and chromatin interactors (e.g. transcription factors) with high sensitivity and resolution.
Request CUT&RUN-seq Quote, Click Here!
Quick Biology CUT&RUN-seq offers many advantages compared to traditional ChIP-seq:
- - Requires fewer cells
- - Lower noise, lower sequencing depths needed
- - Lower cost per sample
- - Faster and simpler workflow
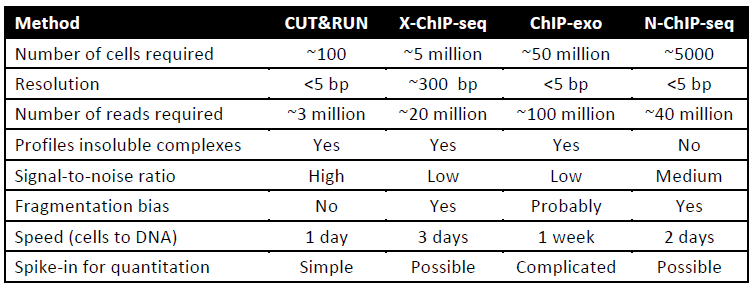
Comparison of CUT&RUN-seq to other ChIP-seq protocols
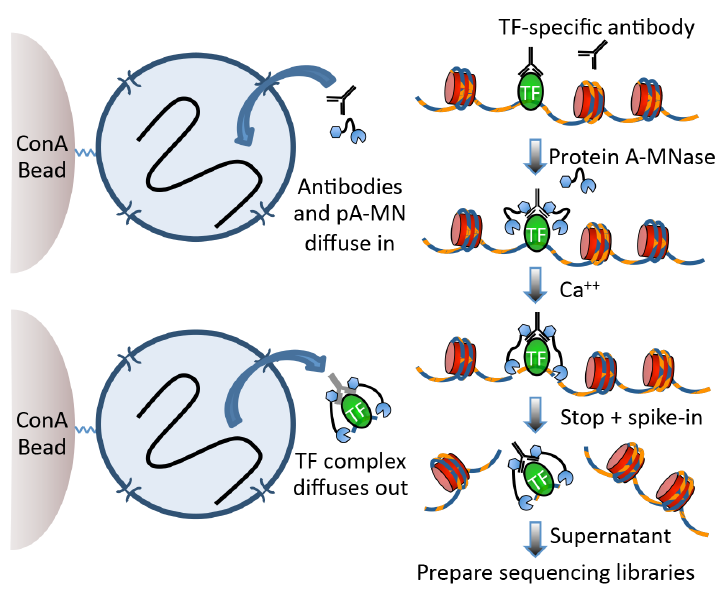
Quick Biology CUT&RUN protocol pipeline. Nuclei attached to magnetic beads can be treated successively with an antibody (or optionally with a primary and secondary antibody) and Protein A-MNase (pA-MN), which diffuse in through the nuclear pores. After Ca++ addition to activating MNase cleavage, fragments are released and diffuse out of the nucleus. DNA extracted from the supernatant is used to prepare libraries for paired-end sequencing.
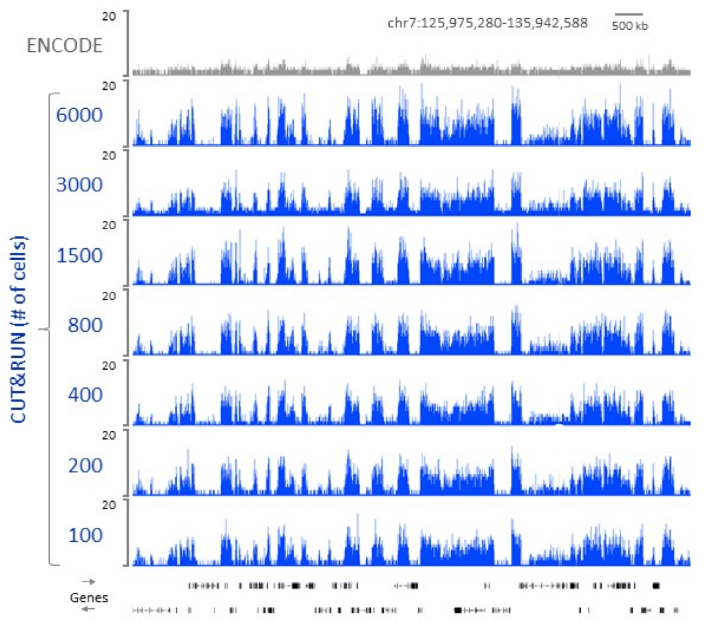
Higher resolution, lower cell number requirement using CUT&RUN protocol.
CUT&RUN of H3K27me3 requires only 100 cells to profile the human Polycomb chromatin landscape. A varying number of K562 cells was used as the starting material for profiling H3K27me3 by CUT&RUN. Following paired-end 2x25 bp Illumina sequencing and removal of duplicates, 7.5 million reads were randomly selected and used to generate bedgraphs representing raw counts, as indicated on the y-axis. For comparison, ENCODE XChIP-seq data (GSM733658) was similarly analyzed.
Bioinformatics for CUT&RUN Sequencing
Standard Analysis:
a.Raw data QC and clean up
b.Alignment to a reference with mapping statistics
c.Peaking calling with or without control samples
d.Gene assignment and peak annotation
e.Pathway Analysis
f. Final project report (HTML) with analysis methods, publication-ready graphics, and references
Advanced Analysis:
h.Motif analysis
i.clustering and visualization
Turnaround time
~4-5 weeks depending on project requirements, one additional week for data analysis.

