2019-11-10 by Quick Biology Inc.
Characterization of host cell response of viral infection is of great interest. Deep insights into molecular signatures of specific cell subsets to viral infection will provide potential antiviral therapies.
In recent Nature communications, Researchers in Germany used Herpesvirus (HSV) infection as a virus-host model, performed single cell RNA-sequencing of HSV-1 infected primary fibroblasts at different infection time points (ref1, Fig 1). Time course of HSV-1 infections experiment related the progression of infection to cell cycle phases and defined a precise temporal order of viral gene expression (Fig.1). Single-cell transcriptome data analysis finally correlated infection resistance with the transcription factor NRF2. In addition, two NRF2 agonists (i.e. Bardoxolone methyl, and Sulforaphane) impaired virus production in cell assays, which strongly suggested that NRF2 activation restricts viral infection (Fig2). This work provides us a vivid example of virus-host molecular interactions by single cell RNA sequencing.
Figure 1: Single-cell RNA-sequencing of HSV-1-infected primary human fibroblasts show cell cycle dependency (ref1). a Infection protocol. In parallel to single-cell RNA-sequencing, cells were harvested for bulk mRNA-sequencing and ICP0 immunofluorescence staining. b ICP0 immunofluorescence staining at 5 hpi. Scale bar: 20 μm. c Global display of scRNA-seq data as tSNE maps. Cells were colored by, from left to right, harvesting time points, cell cycle phase, and the normalized values of the sum of HSV-1 transcripts as a marker for the progression of infection. Cells without HSV-1 transcripts are in light gray. d tSNE maps with cells colored by replicate. e Relative densities of the percentage of viral transcripts (log 10 transformed) per cell for the three time points post infection. f Relative densities of the percentage of viral transcripts per cell (log 10 transformed) for G1 and non-G1 cells for cells harvested at 3 and 5 hpi.
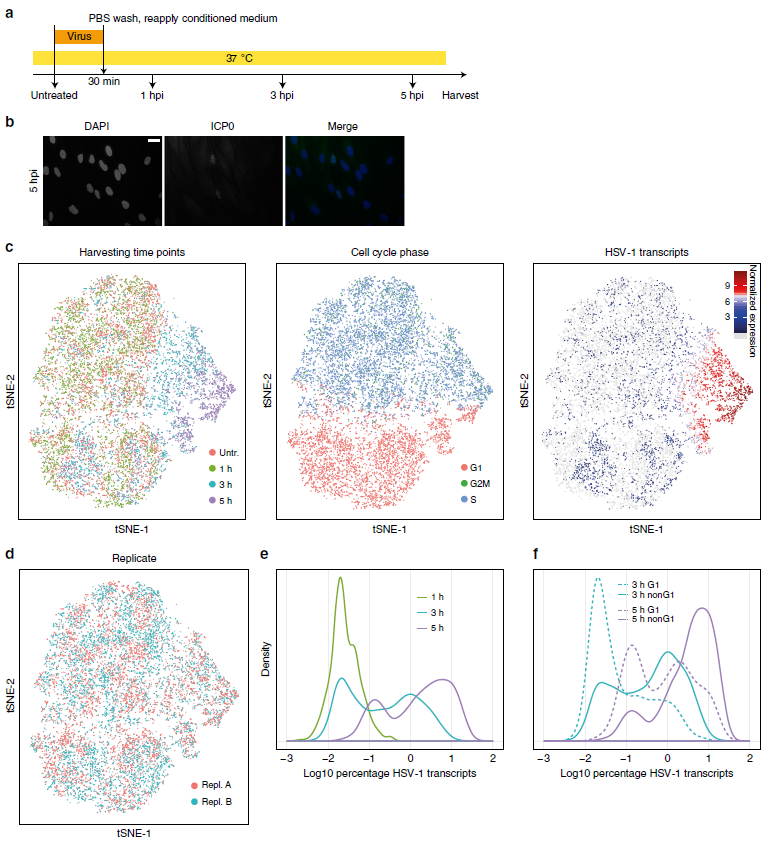
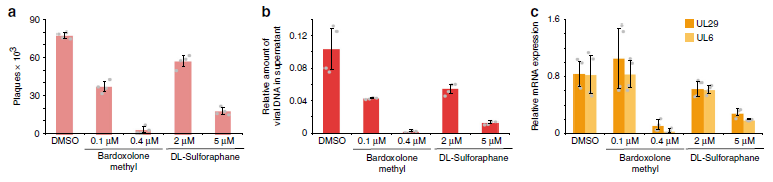
Figure 2: NRF2 activity counteracts HSV-1 infection (ref1). a–c NHDF cells were infected with HSV-1 at an MOI of 1. After the removal of virus inoculum and washing with PBS, conditioned medium supplied with solvent or different concentrations of NRF2 agonists were added. At 16 hpi, virus production was assessed using plaque assays (a), by probing viral DNA in the supernatant using qPCR (b), and by measuring viral mRNAs in the RNA isolated from the cells using RT-qPCR (c). For all panels, bar plots indicate means, and error bars denote standard deviations; the individual measurement values (two each from n = 2 biologically independent samples) are shown as grey dots.
Quick Biology is an expert in single cell, low-input RNA sequencing. Find More at Quick Biology.
Reference:
- 1. Wyler, E. et al. Single-cell RNA-sequencing of herpes simplex virus1-infected cells connects NRF2 activation to an antiviral program. Nat. Commun. 1–14 doi:10.1038/s41467-019-12894-z
