2020-10-26 By Quick Biology
We discussed methods that figure out spatial (location) and temporal resolution in single-cell RNA profiling. ScNT-seq (single-cell metabolically labeled new RNA tagging sequencing) developed by Wu’s Lab as we discussed before, only gives information of newly synthesized transcripts, it lacks the time scale about dynamics of gene expression. To build up a picture of the transcriptional history of a single cell, In Nature Biotechnology, Young scientist Fei Chen at Broad Institute developed RNA timestamps approach for inferring the age of individuals RNAs in RNA profiling by exploiting RNA editing (ref1). Basically, researchers tag RNA with a reporter motif consisting of multiple MS2 binding sites. The fusion protein of adenosine deaminase ADAR2 with MS2 capsid protein will be recruited to MS2 binding sites, meantime, ADAR2 can cause A-to-I edits in tagged RNA. The A-to-I editing level is accumulated by time, allowing the age of the RNA to be inferred with hour-scale accuracy (Fig. 1). They reasoned inferring RNA transcript ages by analyzing the ensemble of timestamped RNAs which was produced by the same promoter. Two keys in this technology:
- a. Engineered ADAR2 which has a catalytic or editing timescale on the order of hours. Such continuous enzymatic activity acting on RNA allows for direct inference of the age of individual RNAs.
- b. The algorithm that infers/calibrate RNA history through RNA edits.
In Summary, RNA timestamps enable us to gather dynamic transcripts in the cells, have great utility for studying the diversity of response to cellular perturbation.
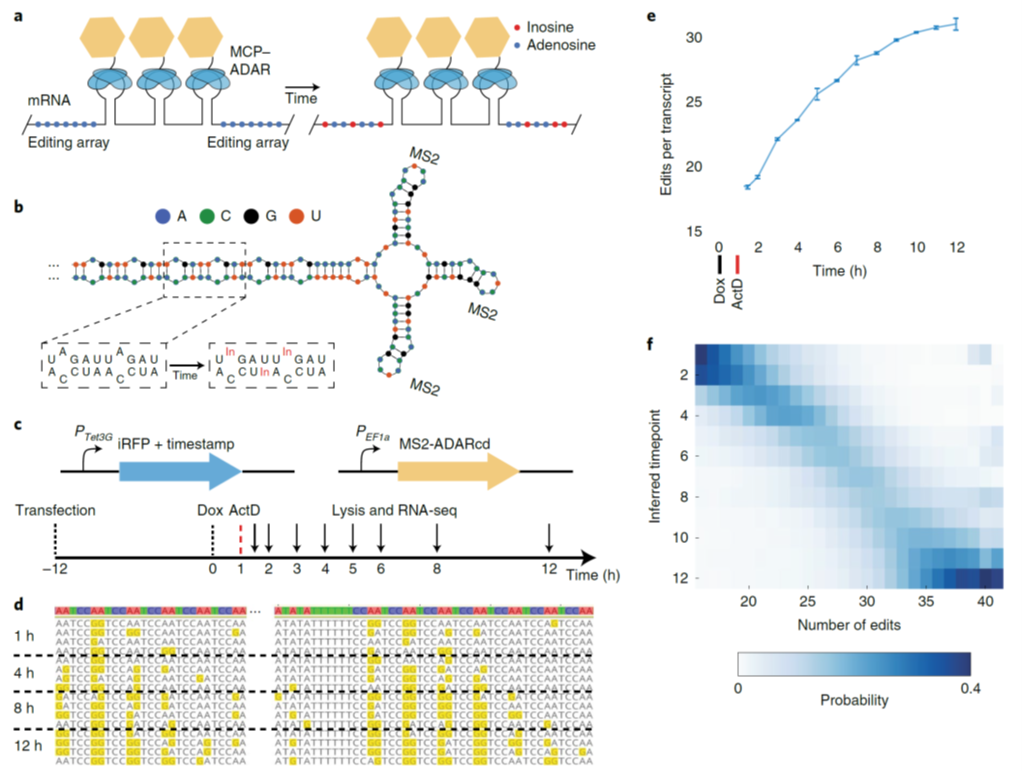
Fig. 1: Encoding of temporal information through RNA edits (image from Ref1). a, Schematic of the timestamps approach, consisting of editing arrays of adenosines (blue dots) and several MS2 step loops in the 3′ UTR of an mRNA. In the presence of an MCP–ADAR fusion (MCP, blue ellipses; ADAR, yellow hexagon), timestamps are edited over time by catalytic conversion of adenosine to inosine (red dots). b, A schematic structure of a portion of one timestamp, showing MS2 stem-loops and the repetitive, double-stranded RNA motif that serves as the editing substrate. c, Schematic representation of the Tet-responsive timestamped mRNA system and experimental timeline. d, Examples of timestamps edited for different amounts of time, showing the accumulation of A-to-I edits. e, The mean number of edits per timestamp observed after editing for different amounts of time. f, The number of edits per timestamp is shown for different specified ages.
Quick Biology is an Expert in single-cell profiling. Find More at Quick Biology.
Ref:
1. Rodriques, S. G. et al. RNA timestamps identify the age of single molecules in RNA sequencing. Nat. Biotechnol. (2020). doi:10.1038/s41587-020-0704-z
