2019-11-03 by Quick Biology Inc.
Population stratification is the presence of a systematic difference in allele frequencies between subpopulations in a population, which possibly due to different ancestry, especially in the context of association studies (https://en.wikipedia.org/wiki/Population_stratification). Precision Medicine must rely on these structured data, analyze the health of patient populations, then make clinical decisions.
Acute myeloid leukemia (AML) is a type of cancer of the blood and bone marrow with excess immature white blood cells. ~ 40% of patients with childhood AML relapse, resulting in a very low overall survival rate at ~70%. Considering the biologic basis differs among pediatric AML patients in United States, Europe, and Japan, in recent Blood advances, the Institute of Physiology and Medicine in Japan aimed to reveal the genetic alternations of all Japanese patients with pediatric AML (ref1). These patients were from the Japanese Pediatric Leukemia/Lymphoma Study Group AML-05 trial. They investigated correlations between genetic aberrations and clinical information (Fig.1). By performing RNA sequencing in 139 of the 369 patients with de novo pediatric AML, they unmasked many gene fusions, and rare gene rearrangements, such as MYB-GATA1, NPM1-MLF1, ETV6-NCOA2, ETV6-CTNNB1, and RUNX1-PRDM16 (Fig.2). Their data indicated that a subset of pediatric AML represents a distinct entity that may be discriminated from their adult counterparts, risk stratification should be reconsidered.
Figure 1: Clinical and genetic profiles of the 369 patients (ref1). (A) Pie chart represented the frequency of each gene alteration. (B) The landscape of 369 de novo pediatric AML patients. Each column indicates 1 patient. *, #Both gene fusions were observed in the same patient. RUNX1-R, RUNX1 rearrangement; NPM1-R, NPM1-rearrangement.
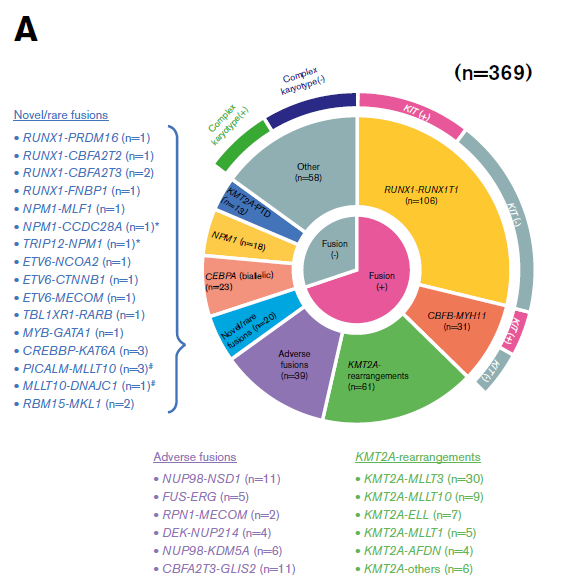
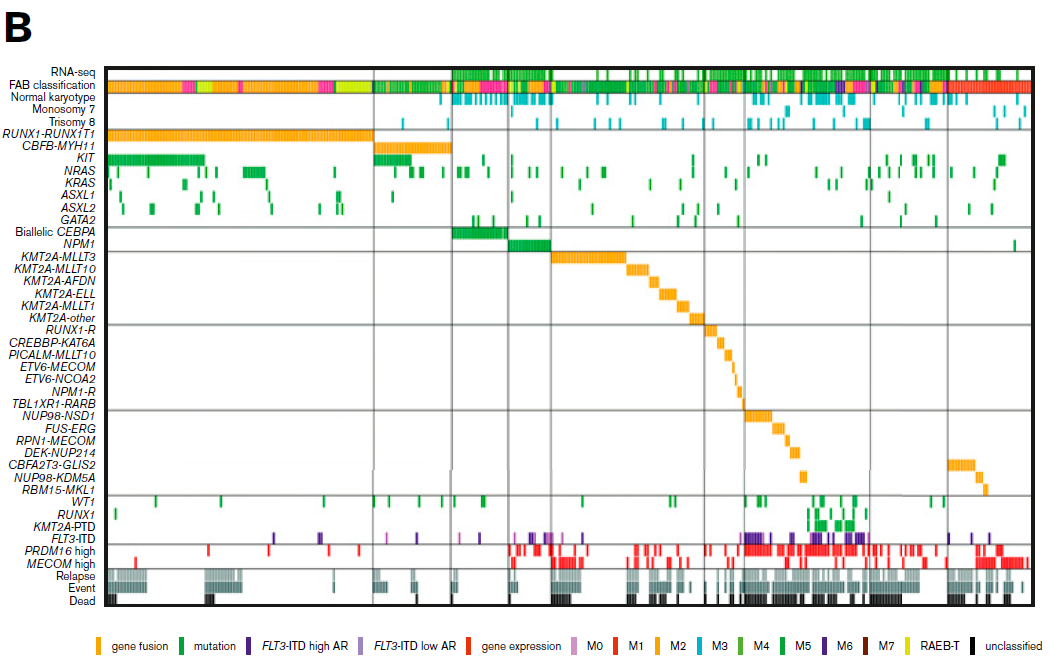
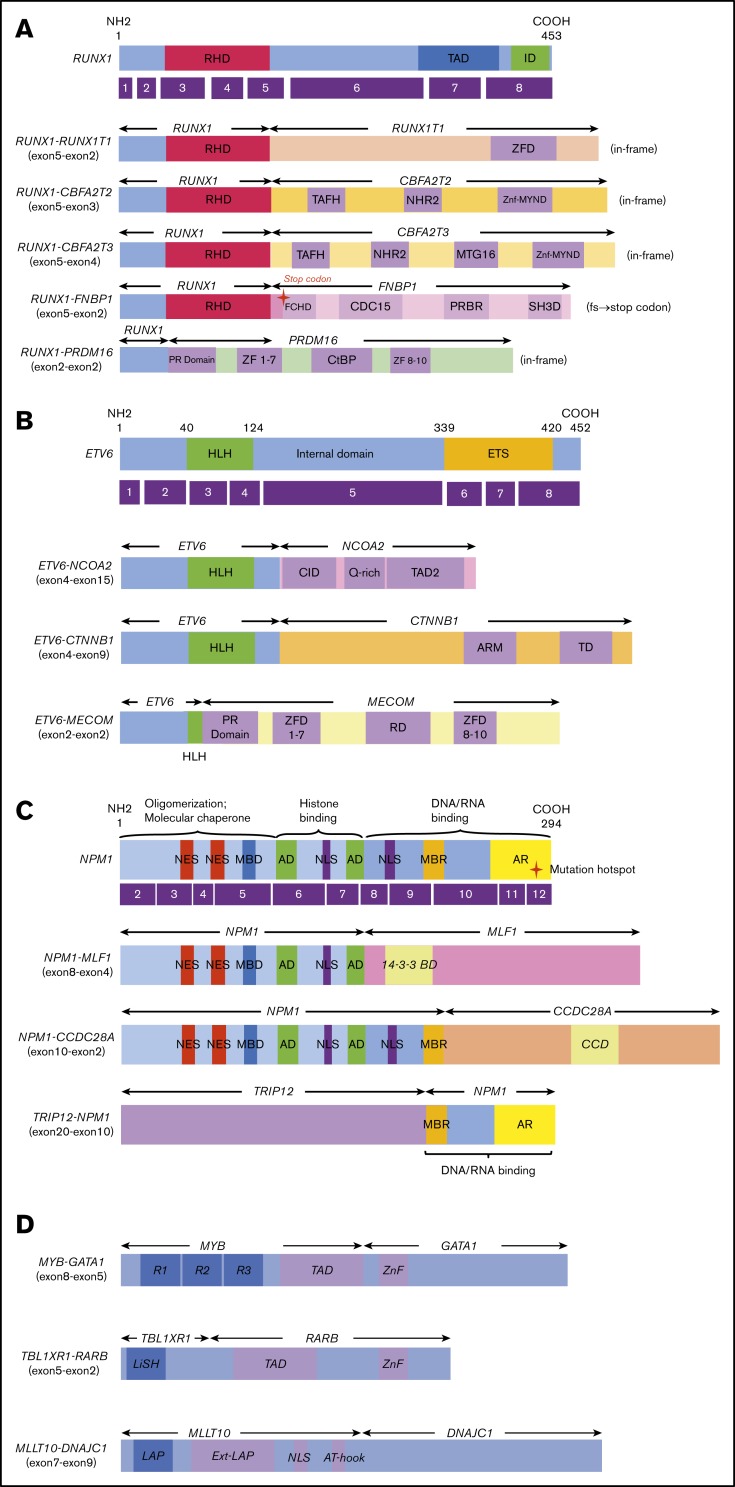
Figure 2: Novel/rare fusion genes identified in patients with de novo pediatric AML. (A-D) The structure of the detected fusion proteins. (A) Five RUNX1 rearrangements. (B) Three ETV6 rearrangements. (C) Three NPM1 rearrangements. (D) Other novel gene fusions. R1, R2, and R3 are 51 6 52 amino acid tandem repeats that comprise the DNA-binding domain Ext–leukemia-associated protein (LAP). 14-3-3 BD, 14-3-3 binding domain; AD, acidic domains; AT-hook, adenine-thymine hook; CCD, coiled-coil domain; CDC15, CDC15 homology region; CID, CBP interaction domain; CtBP, C terminal binding protein; ETS, ETS domain; FCHD, FER-CIP1 homology domain; HLH, helix-loop-helix; ID, transcription inhibition domain; LiSH, Lis homology domain; MBD, metal-binding domain; MBR, moderately basic region; MTG16, myeloid translocation gene 16; NES, nuclear export signal; NHR2, nervy homology region 2; NLS, nuclear localization signal; PRBR, putative r-binding region; RD, runt domain; RHD, runt homolog domain; SH3D, Src homology 3 domain; TAD, transcription activation domain; TAFH, TAF homology; ZF, zinc finger; ZFD, zinc-finger domain; Znf-MYND, MYND zinc-finger 
Quick Biology is an expert in whole transcriptome sequencing and data analysis for identifying fusion genes from RNA-seq data. Find More at Quick Biology.
Reference:
- 1. Shiba, N. et al. Transcriptome analysis offers a comprehensive illustration of the genetic background of pediatric acute myeloid leukemia. Blood advances 3, 1–3 (2019).
