01-19-2021 By Quick Biology
Early detection of causative microorganisms in patients with severe infection is critical to informing clinical interventions and the administration of appropriately targeted antibiotics. The traditional culture-based method is time-consuming (taking days or even weeks), especially some microorganisms are unculturable in a lab setting. PCR tests using conserved 16S ribosomal RNA gene and 28S-ITS PCR regions of bacteria and fungi, respectively have low sensitivity of genus/species level. Specifical clinical tests only focusing on certain pathogens makes it impossible for discovering unknown pathogens as most infectious syndromes present with indistinguishable clinical manifestations.
In Nature Medicine, Dr. Charles Chiu in UCSF-Abbott Viral Diagnostics and Discovery Center (VDDC) describes a simple, rapid, and universal method of pathogen detection by metagenomic next-generation sequencing (mNGS) analysis of cell-free DNA (cfDNA) from a variety of different body fluids, ranging from low-cellularity cerebrospinal fluid (CSF) to purulent fluids with high human host DNA content (ref1). To evaluate their mNGS method sensitivity and precision, they collected 182 samples from 160 patients, performed cultured based pathogen detection, 16S ribosomal RNA, and 28S-ITS PCR assays in parallel (Table 1). To enable pathogen identification within 6h sample-to-answer time, they utilize Nanopore sequencing, a field-deployable, real-time sequencing platform. To minimize the effect of high-rate of sequencing error in the Nanopore sequencing platform, they optimized the sequence and length of sample barcoding/Indexes called “a dual-use barcoding protocol” that is cross-compatible on both nanopore (for the real-time portable setting) and Illumina (for deeper reads coverage and more accuracy in base calling) (Fig.1). This rapid, sequencing-based mNGS test is a promising tool for the diagnosis of unknown infections from body fluids.
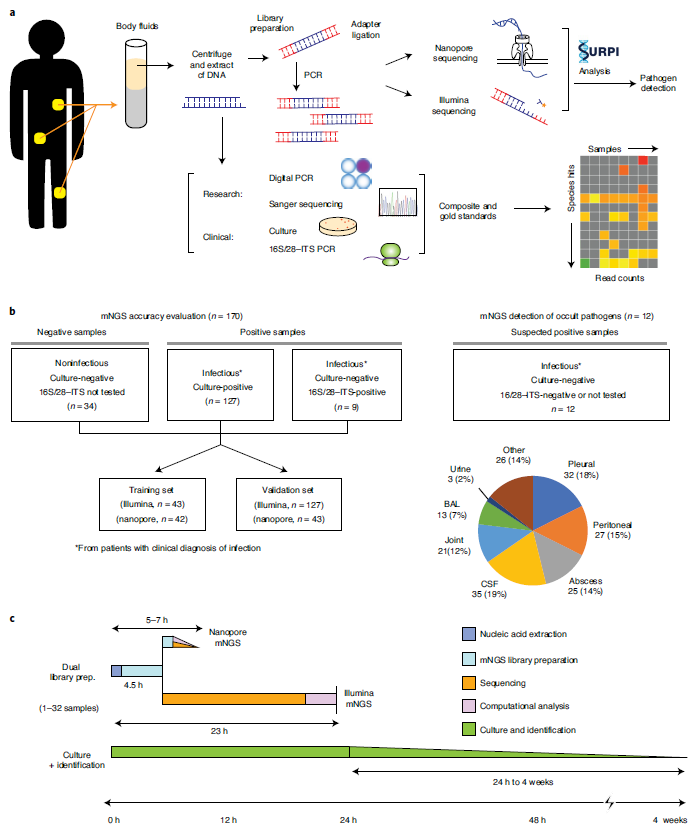
Figure 1: Study workflow and sample distribution. (Ref1). a, Schematic of mNGS body fluid analysis workflow. The clinical gold standard consisted of aggregated results from cultures, bacterial 16S PCR and/or fungal 28S–ITS PCR, while the composite standard also included confirmatory digital PCR with Sanger sequencing and clinical adjudication. For nanopore sequencing in <6 h, 40–60 min are needed for nucleic acid extraction, 2–2.5 h for mNGS library preparation, 1 h for nanopore 1D library preparation and 1 h for nanopore sequencing and analysis. b, Analysis workflow for the 182 body fluid samples included in the study; 170 samples were included in the accuracy assessment while 12 samples collected from patients with a clinical diagnosis of infection but negative microbiological testing were included for mNGS analysis. The pie chart displays the body fluid sample types analyzed in the study. *From patients with clinical diagnosis of infection. c, Timing for mNGS testing relative to culture. Whereas culture-based pathogen identification can take days to weeks, mNGS testing using nanopore or Illumina sequencing platforms has an overall turnaround time of 5–24 h.
Quick Biology can assistant you with both Illumina and nanopore sequencing. Find More at Quick Biology.
See resource:
1. Gu, W. et al. Rapid pathogen detection by metagenomic next-generation sequencing of infected body fluids. Nat. Med. 27, 115-124(2021).
