2019-10-20 by Quick Biology Inc.
Unlike next-generation sequencing (specifically, Illumina Sequencing by Synthesis Technology), new third-generation sequencing is being developed to overcome limitations such as amplification biases, short reads (reads size < 300 bp).
Oxford Nanopore Technology uses nanopore-based sequencing to detect changes in current as a single strand of nucleic acid sequence transverses a pore protein (Fig.1). Single nucleic acid Modification of DNA or RNA will change the current. By deconvoluting these electrical signals, the specific nucleotide modification can be reconstructed. In recent RNA, Yeo Lab at UCSD developed a Random Forest classifier trained using experimentally detected m6A sites----MINES, this software assigned m6A status to over 13,000 previously unannotated DRACH sites (Fig.2) and identified over 40,000 sites with isoform-level resolution in a human cell line (Fig.3 as an isoform-level example). Further cross-validation or other software/methods comparison is highly recommended for the current m6A detection field.
Figure 1: Nanopore sequencing technology. A protein nanopore is set in an electrically resistant polymer membrane. An ionic current is passed through the nanopore by setting a voltage across this membrane. If an analyte passes through the pore or near its aperture, this event creates a characteristic disruption in current (as shown in the diagram below). Measurement of that current makes it possible to identify the molecule in question. In the figure, a strand of DNA is passed through a nanopore. The current is changed as the bases G, A, T, and C pass through the pore in different combinations (https://nanoporetech.com/how-it-works).
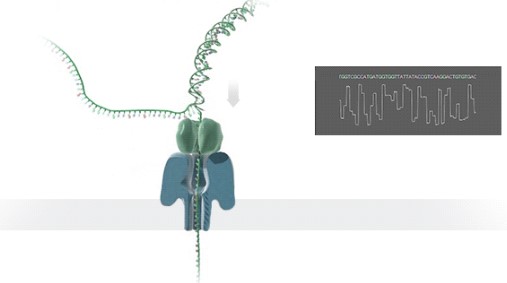
Figure 2: MINEs predicts m6A within DRACH motifs. (Left panel) ROC curve for GGACT motif from the final model. (Right panel) Venn diagram for the prediction of AGACT, GGACA, GGACC, and GGACT sites. CLIP sites represent data withheld from training for testing purposes (ref1).
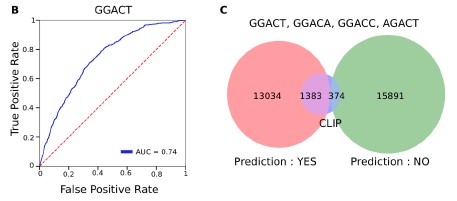
Figure 3: MINES provides m6A isoform-level resolution. (ref1).
Quick Biology is an expert in Next Generation Sequencing Platform. Contact us if you have any questions about your study design or sequencing platform. Find More at Quick Biology.
Reference:
