2019-10-13 by Quick Biology Inc.
Tumor heterogeneity is the critical factor harboring resistance mechanisms. During drug treatment, the evolutionary pressure drives outgrowth of distinct tumor subclones, which finally results in acquired resistance of current targeted therapies.
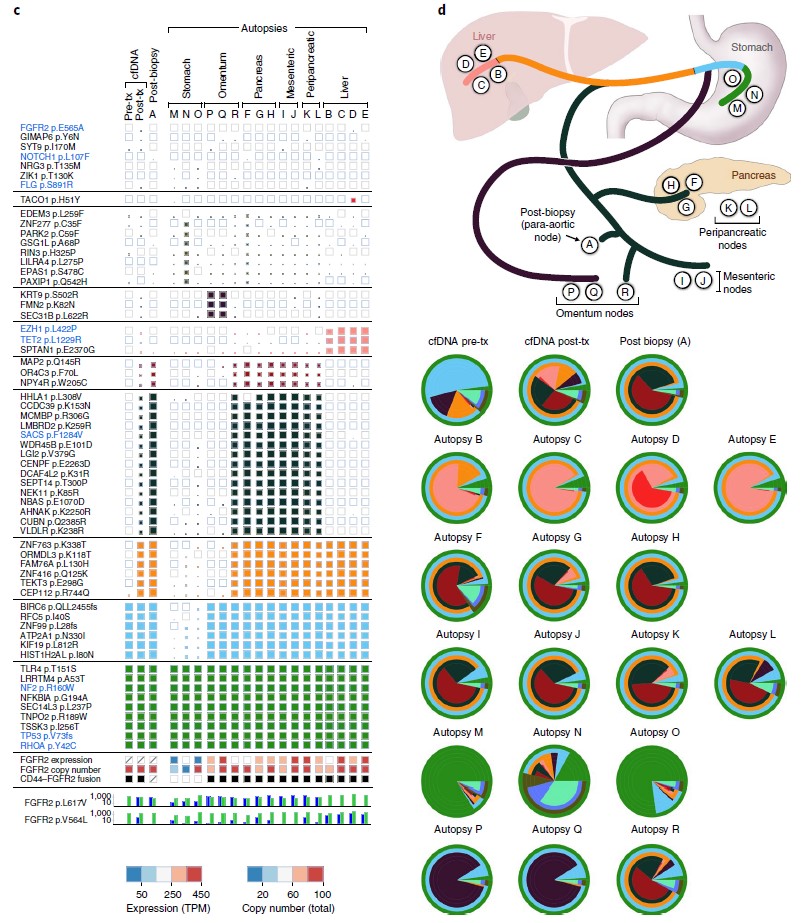
As we discussed before (https://www.quickbiology.com/news/incorporation-liquid-biopsy-and-ngs-powerful-strategy-precision-medicine), liquid biopsy (specifically, cell-free DNA) can be used for capturing acquired tumor mutations. In recent Nature Medicine (ref1), researchers directly compared the effectiveness of cfDNA versus standard single-lesion tumor biopsies in a cohort of 42 patients with gastrointestinal cancers. Interestingly, cfDNA was more sensitive, it identified clinically relevant sequence mutations, which was not found in the matched tumor biopsy in 78% cases (Fig.1). Time-course serial cfDNA, tumor biopsies' WES analysis elucidated substantial geographic and evolutionary differences across lesions (Fig.2). These findings highlight the potential advantages of cfDNA (not only cfDNA is easily obtained, minimal pain/risk) over tissue biopsy in the setting of acquired resistance alterations.
Figure 1: Identification of mutations in liquid versus tumor biopsy. Comparison of specific resistance alterations identified in plasma cfDNA (n = 42) versus tumor biopsy (n = 23) for each patient. Patients are grouped according to tumor type (n = 3; CRC, gastroesophageal (GE) or biliary) and molecular subtype (n = 5; FGFR2 fusion (FGFR2), MET amplification (MET), HER2 amplification (HER2), BRAFV600E or RAS wild type (RAS WT)). (ref1)
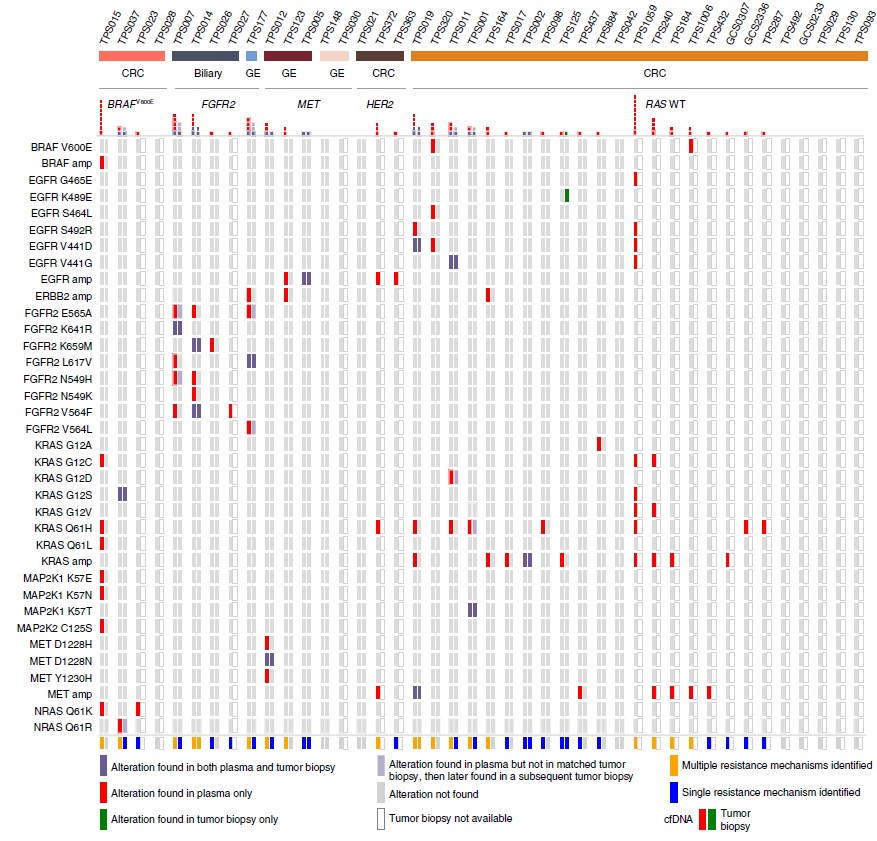
Figure 2: Serial liquid biopsy and autopsy in a patient with FGFR2 fusion-positive gastric cancer. c, Clonal and subclonal alterations detected in each plasma (n = 2), tumor biopsy (n = 1) and autopsy specimen (n = 17). d, Diagram of the locations of the autopsy specimens included in the analysis, along with the likely clonal migration patterns across lesions. Layered pie charts represent the estimated clonal composition of each specimen, with the color of each subclone matching the respective gene color in the branches of the phylogenetic tree. Tx, treatment. (ref1)
Quick Biology is an expert in whole-exome sequencing and cfDNA sequencing. Are you ready to sequence your samples of interest? Find More at Quick Biology.
Reference:
- 1. Parikh, A. R. et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat. Med. 25, 1415–1421 (2019).
