2020-09-14 by Quick Biology Inc.
We previously introduced Oxford Nanopore sequencing, a third-generation sequencing that read the nucleotide sequences at a single-molecule level. It has many advantages than the current Illumina sequencing platform (see News), however, its output and reads accuracy so far is far inferior to that of Illumina. Quick Biology believes the products of Illumina and Oxford Nanopore have different applications, but both will occupy a certain share in the current high throughput sequencing market to meet different needs. Sequencing native RNA directly is a proprietary technology in third-generation sequencing. However, Oxford Nanopore has not released a protocol for indexing direct RNA sequencing samples (Fig.1). Combining multiple samples on the same flow cell, which is a common strategy in the Illumina Hi-Seq series, can greatly improve the cost-effectiveness.
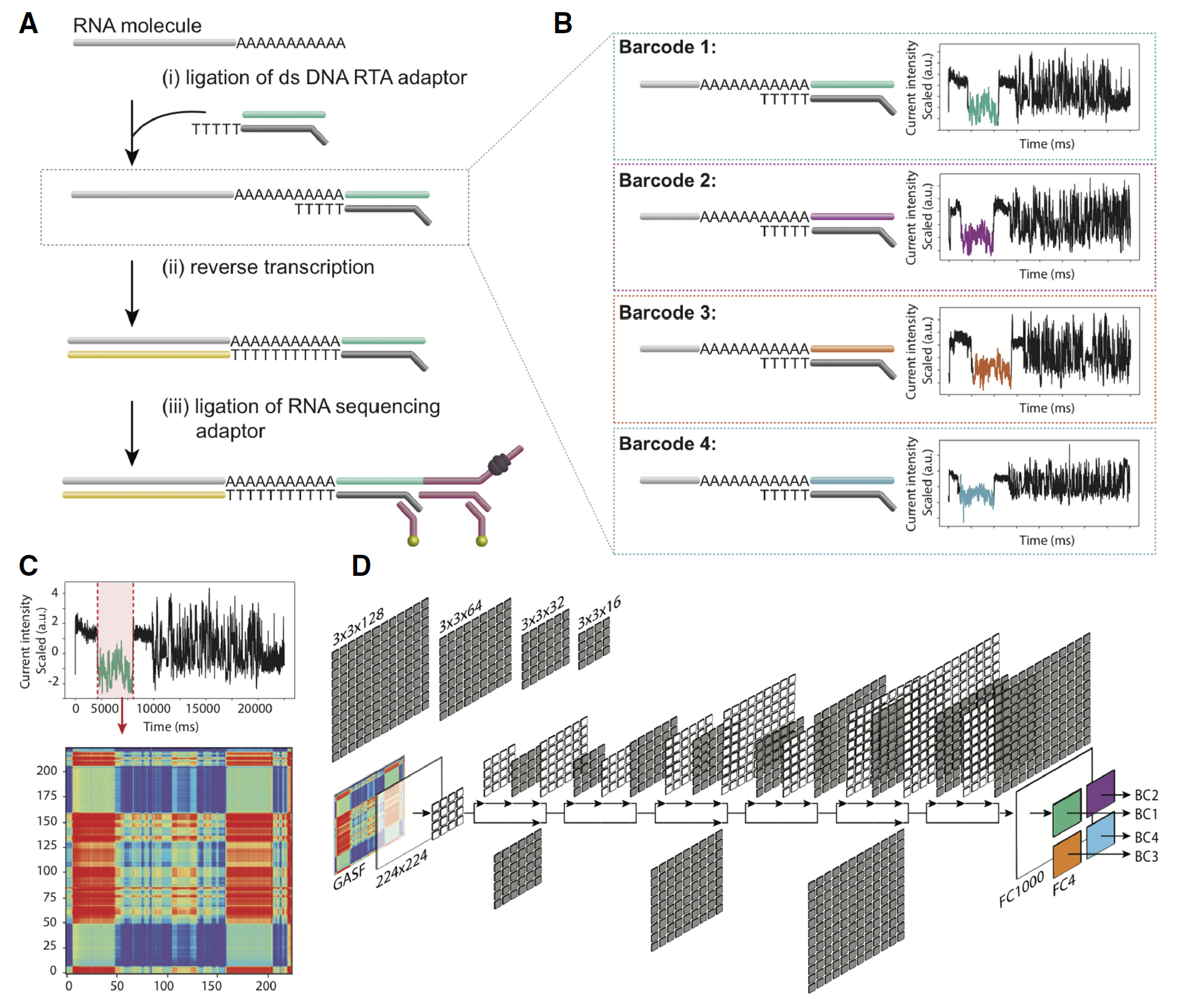
In Genome Biology, researchers at Garvan Institute of Medical Research publishes the first experimental protocol and deep learning method called DeePlexiCon to barcode and demultiplex direct RNA nanopore sequencing data (Fig.2). Briefly, three custom DNA barcode adapters by shuffling the double-stranded sequence of the default ONT RTA (Reverse Transcription Adapter). The three custom barcodes, together with the standard ONT RTA, were individually ligated to four distinct in vitro–transcribed RNA sequences. Electric current for a certain barcode is extracted, and segmented and followed by classification using a deep learning algorithm. DeePlexiCon can classify 93% of reads with 95.1% accuracy or 60% of reads with 99.9% accuracy, the authors provide a novel experimental protocol and associated algorithm to barcode and demultiplex direct RNA Nanopore sequencing.
Fig. 1: Nanopore product line for RNA sequencing (screenshot from ref1, Nanopore product brochure).
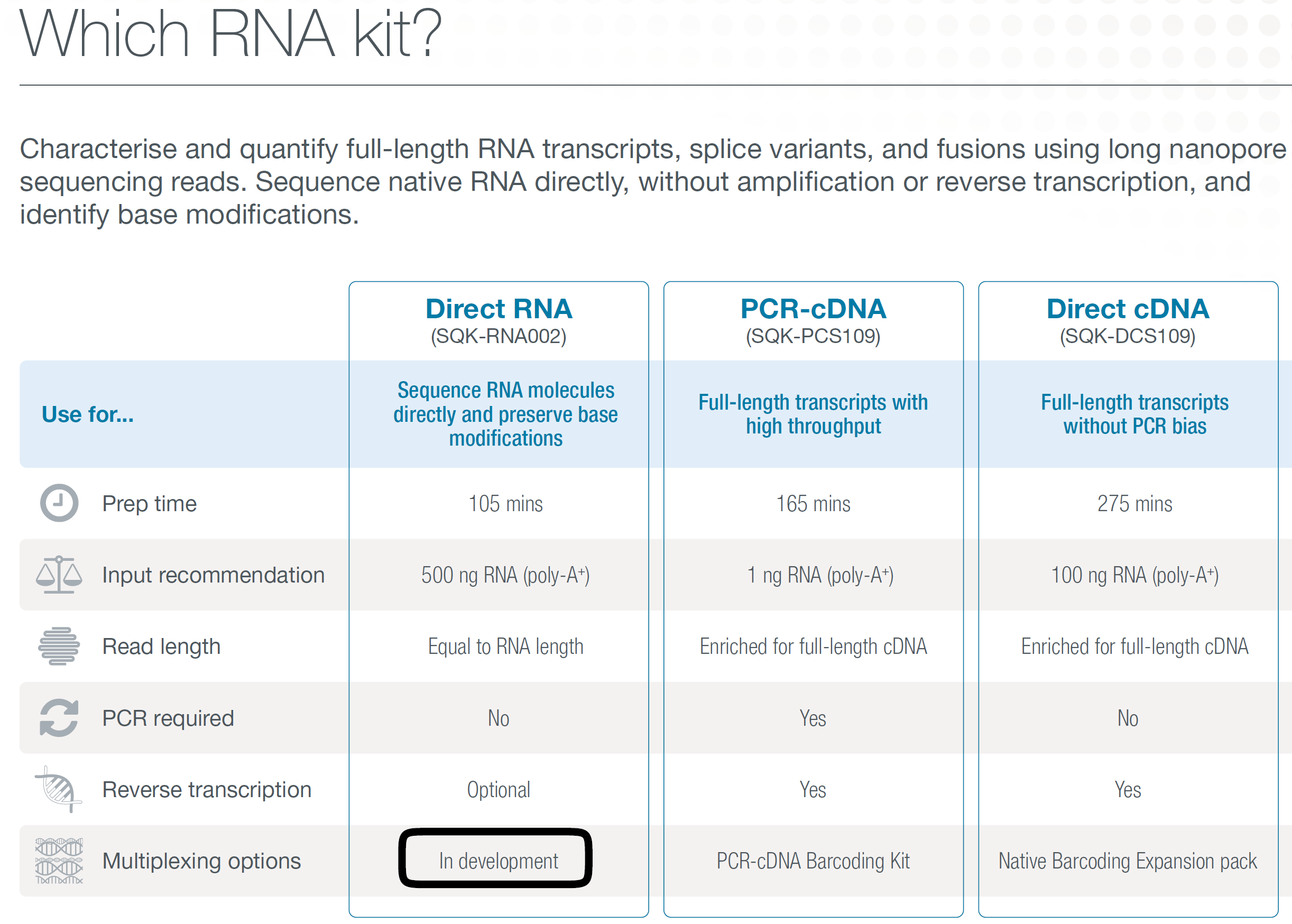
Fig. 2: Schematic overview of the direct RNA barcoding and demultiplexing strategy. (A) Overview of Oxford Nanopore library preparation protocol for native RNA sequencing. (B) Adaptation of A to include custom DNA barcodes. (C) Barcode segmentation and transformation, where the electric current associated with a barcode adapter (highlighted in red) is extracted and converted into an image using GASF transformation. (D) Deep learning is used to classify the segmented and GASF-transformed squiggle signals into their corresponding bins, without the need of base-calling the underlying sequence. The convolution architecture of the final residual neural network classifier (ResNet-20) described in this work: FC = fully connected layer. (from ref2).
Quick Biology offers Nanopore sequencing for your project. Find out More at Quick Biology.
Ref:
1. Nanopore product brochure (Version Jan 2020): https://nanoporetech.com/sites/default/files/s3/literature/product-broch...
2. Smith MA, Ersavas T, Ferguson JM et al. (2020) Molecular barcoding of native RNAs using nanopore sequencing and deep learning. Genome Res [online ahead of print].

