2020-11-9 By Quick Biology
CRISPR is a promising tool for correcting genetic defects to prevent or cure genetic disease. However, gene therapy targeting DNA is irreversible, is potentially damaging to germline cells. Therefore, it has been of great interest to target RNA in somatic cells while avoiding permanent off-target genomic edits.
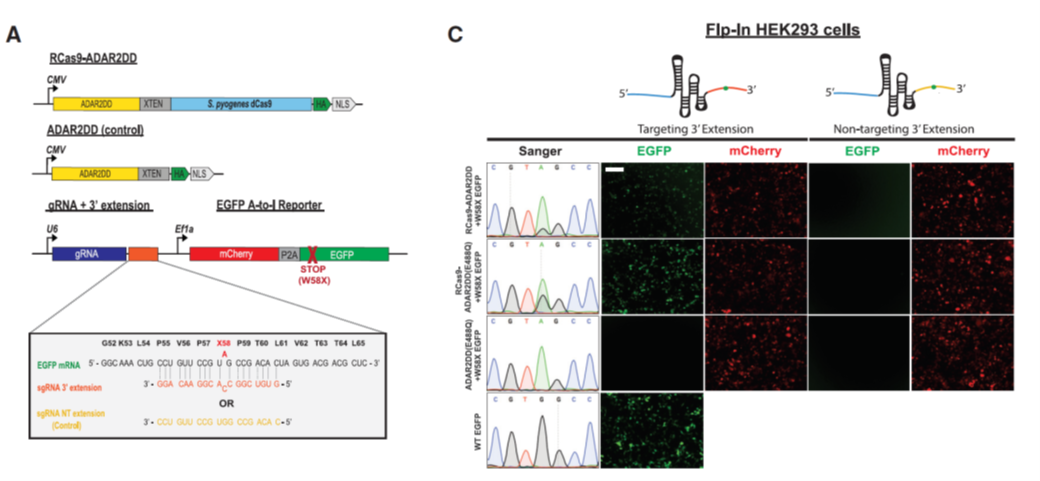
In Cell Reports, Yeo Lab in UC, San Diego developed a nuclease-dead RNA-targeting CRISPR-Cas systems (ref1). They synthesized Streptococcus pyogenes Cas9-ADAR2 fusion protein. Using 3’ modified guide RNA (gDNA), modular Cas9 part in fusion protein can act as an RNA-aptamer-binding protein, specifically be recruited in target site in certain transcripts; Meanwhile, modular ADAR2 part carrying deaminase domain can perform A-to-I RNA editing (Fig. 1). To evaluate this system, they also used a W58X EGFP reporter co-transfected into cells. Only with A-to-I RNA editing, the reporter can have a correct EGFP ORF, give fluorescent signals (Fig. 2). By comparing other RNA-targeting Cas platforms and using RNA whole transcriptome sequencing, they found these strategies have distinct transcriptome-wide off-target edits.
Highlights:
Yeo Lab’s Cas9-ADAR system targets exogenous and endogenous transcripts in a spacer-independent manner, giving more flexibility for certain gene site manipulation.
Figure 1: Graphical abstract of CRISPR-Cas-mediated RNA editing system. (ref1)
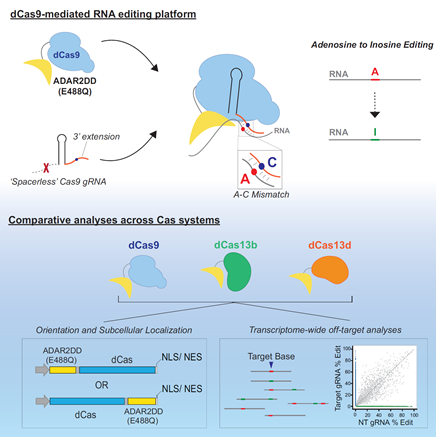
Figure 2: Editing Cellular Transcripts by using RCas9-ADAR2DD (ref1). (Left) Schematic of RCas9-ADAR2 deaminase domain (DD) fusion constructs and modified guide RNA (gRNA) constructs fused to an W58X EGFP reporter. (Right) Sanger electropherograms and representative fluorescent images for EGFP+ cells measuring successful RNA editing of the W58X reporter in an RCas9-ADAR2DD, hyperactive mutant (RCas9-ADAR2DD(E488Q)) or ADAR2DD(E488Q) control expressing Flp-In T-REx 293 lines along with either a targeting or a reverse complement non-targeting (NT) 30 extension (30ex) sequence attached to the gRNA following doxycycline induction. As a transfection control, cells were also imaged for mCherry+ signal. Scale bar, 500 mm.
Quick Biology is Expert in High throughput sequencing screening. Find More at Quick Biology.
See resource:
1. Marina, R. J., Brannan, K. W., Dong, K. D., Yee, B. A. & Yeo, G. W. Evaluation of Engineered CRISPR-Cas-Mediated Systems for Site-Specific RNA Editing. Cell Rep. 33, 108350 (2020).
