2019-09-29 by Quick Biology Inc.
DNA methylation and demethylation are tightly regulated, they shape chromatin accessibility and control gene expression during development. The Ten-Eleven Translocation (TET) genes play important role in DNA methylation and demethylation homeostasis.
In recent Nature communications, researchers in Texas A&M University used Tet2, Tet3 mice deletion mouse model, analyzed DNA methylation dynamics and organization during early heart development. In this comprehensive research project, the authors first performed whole-genome bisulfite sequencing (WGBS, for 5mC profiling) and CMS-IP-seq (for 5hmC profiling) in both human and mouse embryonic heart tissues. After finding impaired 5hmC, they performed RNA-seq to figure out which genes are dysregulated after Tet2/Tet3 deletion. To further verify which specific cell types (i.e. myocardium, epicardium, endocardium or fibroblasts) dysfunction, they performed single cell RNA sequencing. To correlate epigenetic modification with genome-wide chromatin accessibility, they performed ATAC-seq assay. This study provides a vivid physiological role of Tet/YY1 in regulating epigenetic dynamics in heart development.
Figure 1: Genome browser view examples of decreased transcription (blue, RNA-seq), chromatin accessibility (green, ATAC-seq) and 5hmC (red, CMS-IP) enrichment at cardiac-specific gene loci (Tnnt2 and Nppa) in Tet2/3-DKO E12.5 heart tissues compared with that in control group. (ref1)
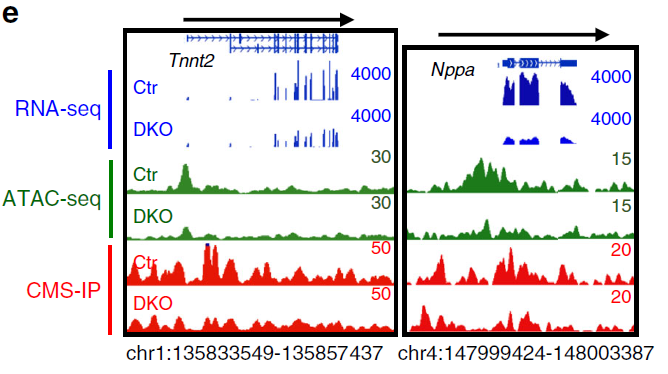
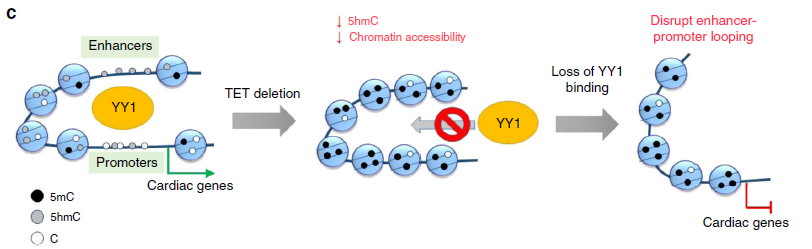
Figure 2: Tet-Chromatin regulation model. Tet protein mediated DNA hydroxymethylation regulates chromatin accessibility and subsequently safeguards the binding of key cardiac development-associated transcription factors, such as YY1, to their target regions in the genome to maintain proper long-distance interactions (enhancer-promoter looping). Deletion of Tet protein, with consequent 5hmC loss, could reduce chromatin accessibility to compromise YY1 binding to its genomic targets, thereby affecting long distance interactions to perturb the transcriptional networks underlying normal cardiac development.
Quick Biology is an expert in Next Generation Sequencing such as WGBS, ATAC-seq, RNA-seq, scRNA-seq used in the research as shown above. Are you ready to sequence your samples of interest? Find More at Quick Biology.
Reference:
- Fang, S. et al. Tet inactivation disrupts YY1 binding and long-range chromatin interactions during embryonic heart development. Nat. Commun. 10, 1–18 (2019).
