2020-06-29 by Quick Biology Inc.
Orchestrated gene expression is critical in cell differentiation and organ development. Gene transcripts can be controlled in different layers including but not limited to: a) Temporal and spatial expressed transcription factor can binding to certain genes’ promoter region, trigger their expression; b) mRNA-alternative splicing coupled to nonsense-mediated decay of transcripts ensure no toxic truncated protein product; c) 5’ UTR (Untranslated Region) of transcript contains various regulatory elements, play a major role in translation initiation; d) 3’ UTR carrying lots of miRNA binding sites plays a role in mRNAs’ half-life, mRNA stability, in final protein production.
In Nature communications, Tian Bin’s group (Now moved to the Wistar Institute, https://wistar.org/our-scientists/bin-tian) discovered widespread 3’ UTR shortening in the differentiation of syncytiotrophoblast (SCT), a cell type critical for hormone production and secretion during pregnancy (Ref1). By analyzing alternative polyadenylation (APA) in previously RNA-seq data, the authors found that differentiation of hESCs (human embryonic stem cells) into trophoblast (TB) showed 3’ UTR shortening, while differentiation into other lineages led to 3’ UTR lengthening (Fig.1). To study TBs lineage in details, authors performed a 3’READs-seq, a method to specifically interrogate 3’ ends of poly(A)+ RNAs, they found that in differentiation, cells prefer choosing proximal polyadenylation site (pPAS), intronic polyadenylation site (IPA), resulting in a widespread 3’ UTR shortening (Fig.2). Further single cell profiling indicated that this alternative polyadenylation regulation correlates with syncytiotrophoblast formation. In summary, researchers reported the unexpected transcript shortening during the differentiation of SCT(Fig.3).
Figure 1: Trophoblast differentiation displays a unique APA profile. a. Schematic showing differentiation of human embryonic stem cells (hESCs, H1 line) into four lineages. b Schematic of 3ʹUTR APA analysis using RNA-seq data. RNA-seq reads in 3ʹUTRs are divided into constitutive UTR (cUTR) and alternative UTR (aUTR) groups based on the first and last PASs, as indicated. 3ʹUTR length change for a gene is indicated by relative expression difference (RED) value between two samples or sample groups, as shown in the graph. RED is calculated as indicated. c Median REDs of four differentiation lineages as compared to hESC. A total of 8,712 genes were used for analysis. d Boxplots showing expression changes of proliferation genes (left; 350 genes; defined by Sandberg et al.) and development genes (right; 74 genes; defined by Ji et al.) in TB vs. hESC. P-values (Wilcoxon test) based on comparisons with other genes are indicated (Ref1).
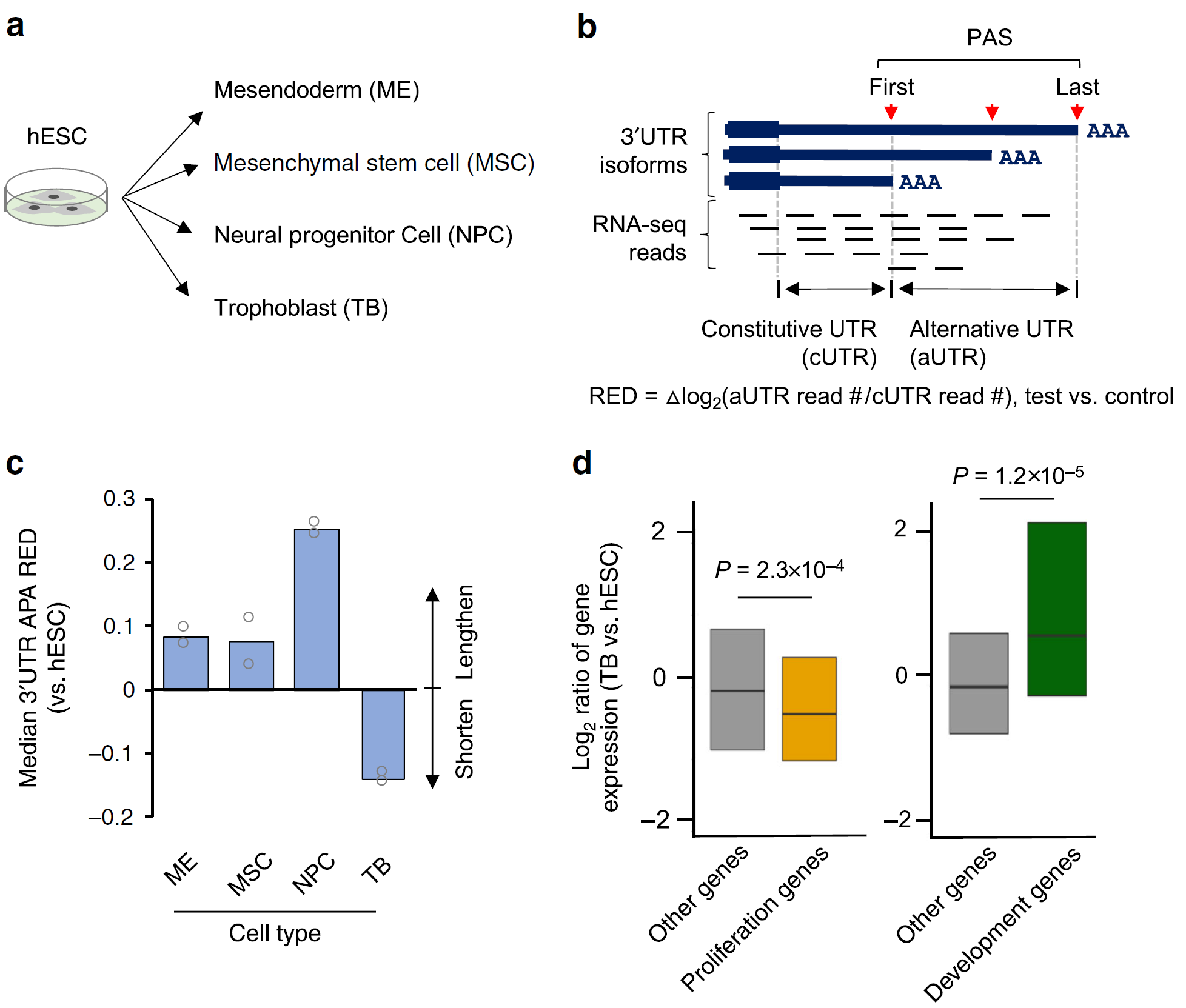
Figure 2: Widespread 3’ UTR shortening and IPA activation in trophoblast differentiation. a Schematic of 3ʹREADS experiment using hESCs (H9 line) treated with BMP4 for TB differentiation. b Schematic of 3ʹUTR APA analysis using 3ʹREADS data. Two representative 3ʹUTR isoforms are selected, named proximal and distal PAS (pPAS and dPAS) isoforms, respectively. RED is calculated as indicated. c Scatter plot showing 3ʹUTR APA changes. Each dot is a gene with two selected 3ʹUTR isoforms. The numbers of genes showing significantly lengthened 3ʹUTRs (red) or shortened 3ʹUTRs (blue) are indicated, and so is the ratio of these two numbers. Significance here and other panels of this figure is based on P < 0.05 (Fisher’s exact test) and > 20% change of expression ratio of the two isoforms. d Relationship between aUTR size and degree of 3ʹUTR size regulation (represented by RED). Genes are grouped into five similarly sized bins (about 500 genes each) based on their aUTR length. aUTR size range for each bin is shown in a table. Data are presented as median value ± SEM. P-value (Wilcoxon test) comparing Bins 1 and 5 is indicated. e Schematic of intronic polyadenylation (IPA) isoforms and 3ʹ terminal exon polyadenylation (TPA) isoforms. f Scatter plot showing IPA changes. Each dot is a gene with expression of both IPA and TPA isoforms. The numbers of genes with significant IPA suppression (red) or IPA activation (blue) are indicated, and so is their ratio (Ref1).
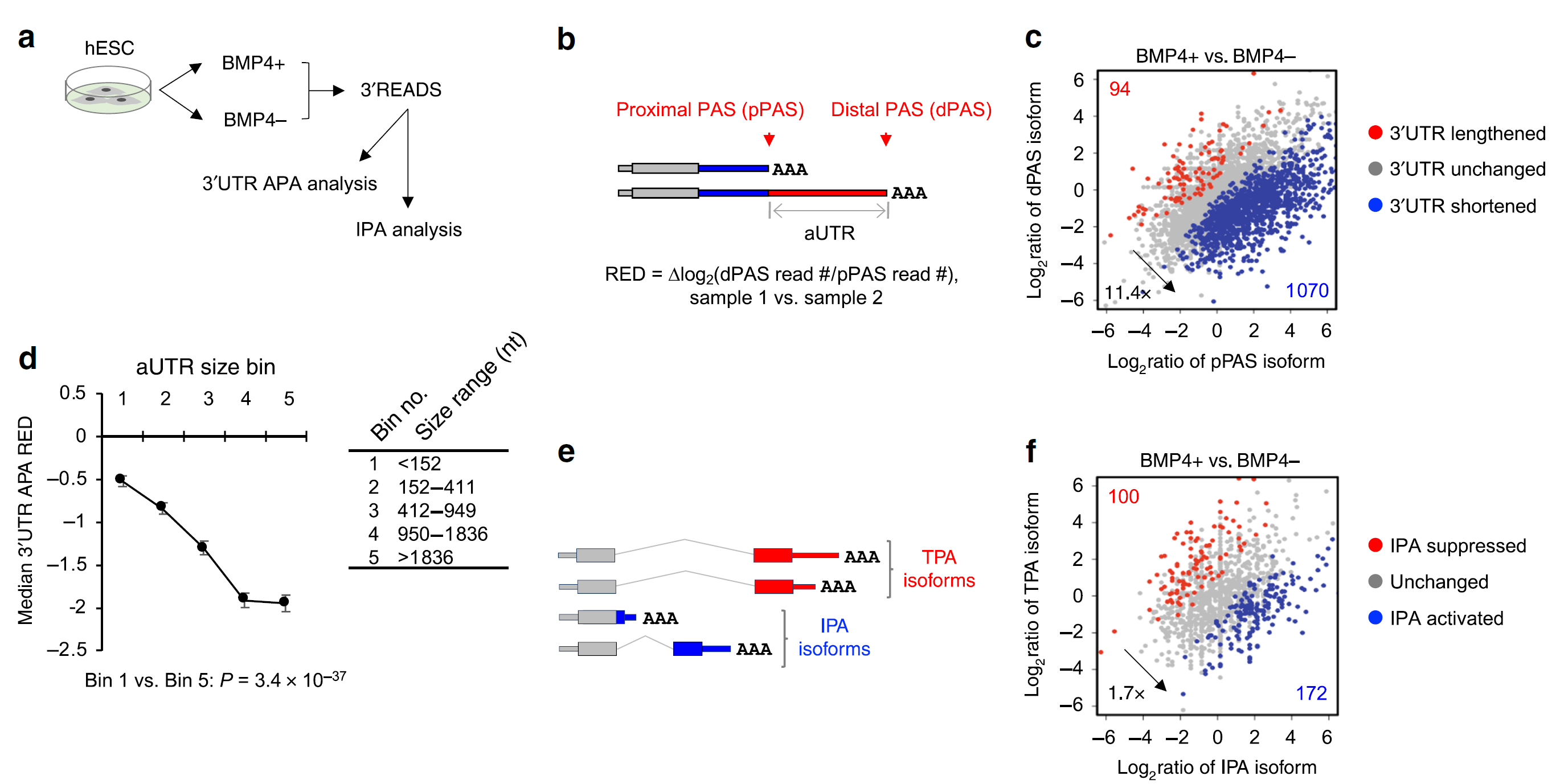
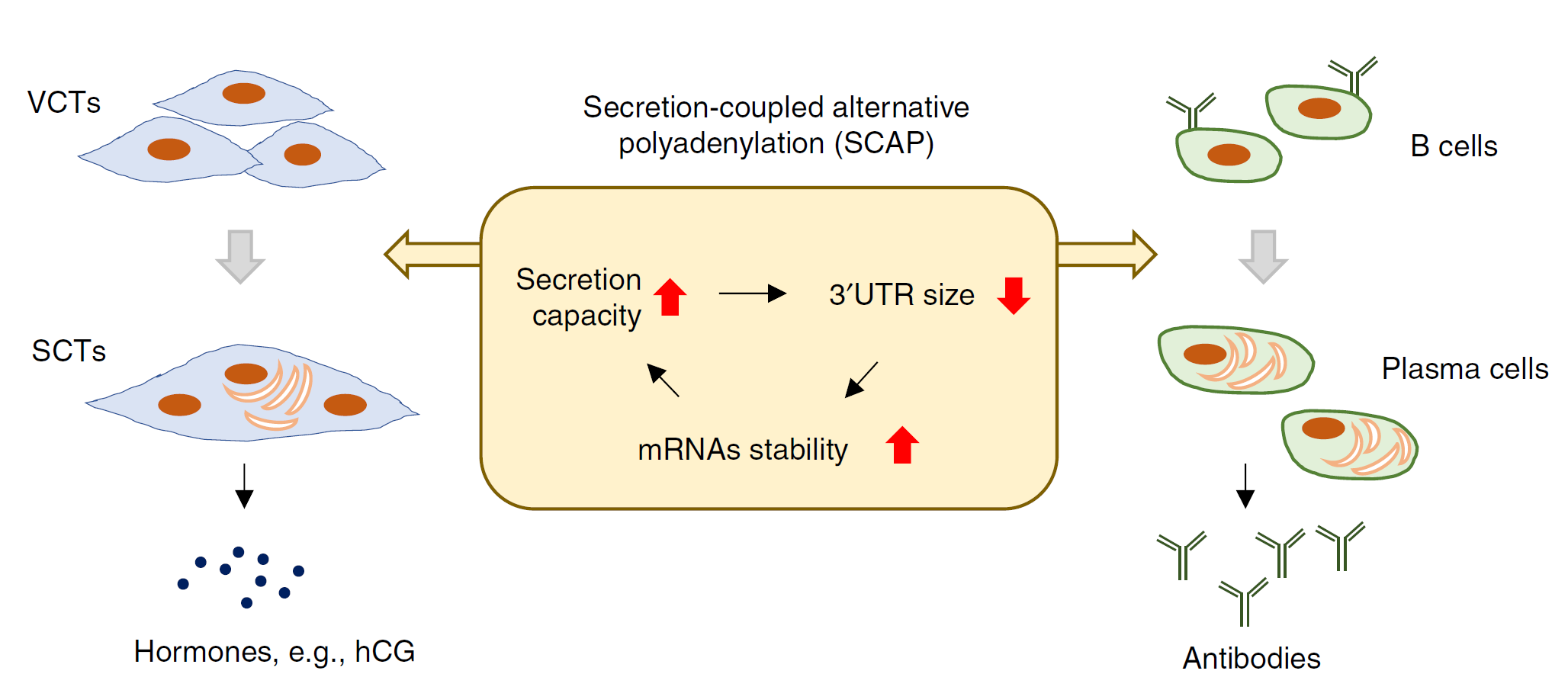
Figure 3: Secretion-coupled APA in the differentiation of secretory cells.
Quick Biology is an expert in NGS applications, and we can help you with the 3' UTR sequencing project. Find More at Quick Biology.
Ref:
1. Cheng, L. C. et al. Widespread transcript shortening through alternative polyadenylation in secretory cell differentiation. Nat. Commun. 11, 1–14 (2020).
